Pathogenic microorganism genome database and establishment method thereof
A technique for pathogenic microorganisms and establishment methods, which is applied in the field of pathogenic microorganism genome databases and its establishment, can solve problems such as increased analysis costs, false negative test results, and fast analysis timeliness, so as to reduce the demand for analysis and computing resources and improve detection accuracy , The effect of shortening the analysis time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] A pathogenic microorganism genome database is established by the following method:
[0049] 1. Data acquisition
[0050] Download bacterial genome data from PATRIC as follows:
[0051] The full name of PATRIC is the American Pathogenic Microorganism Resource Integration Center. The website contains most of the currently known bacterial pathogenic microorganism genome data, and all collected bacterial genome data can be downloaded from its ftp server.
[0052] On the PATRIC website ftp server (ftp: / / ftp.patricbrc.org / ), all genome data classified as archaea and bacteria were downloaded, and the corresponding genome information statistics file PATRIC_genome.txt was downloaded, which contained genome information of 227,577 strains in total.
[0053] 2. Strain Genome Screening
[0054] According to the header information of the file, select the genomes whose "Public" column is "True", "Genome Status" is "Complete", and "Genome Quality" is "Good". After screening, 13537 ge...
Embodiment 2
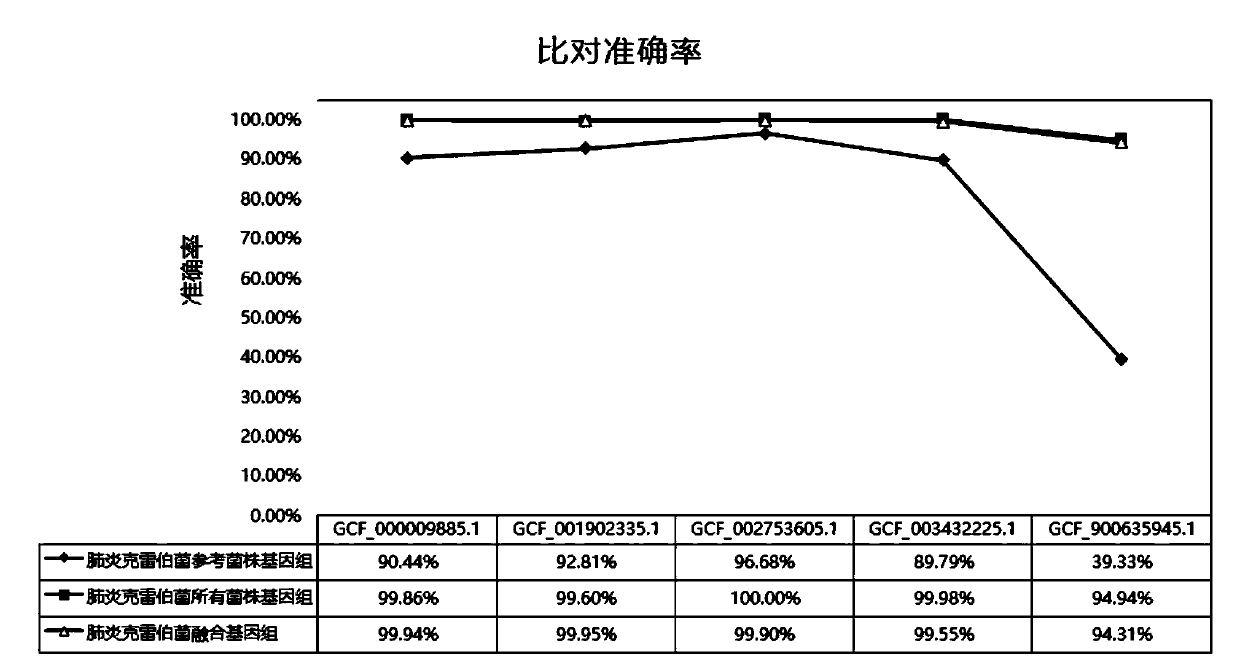
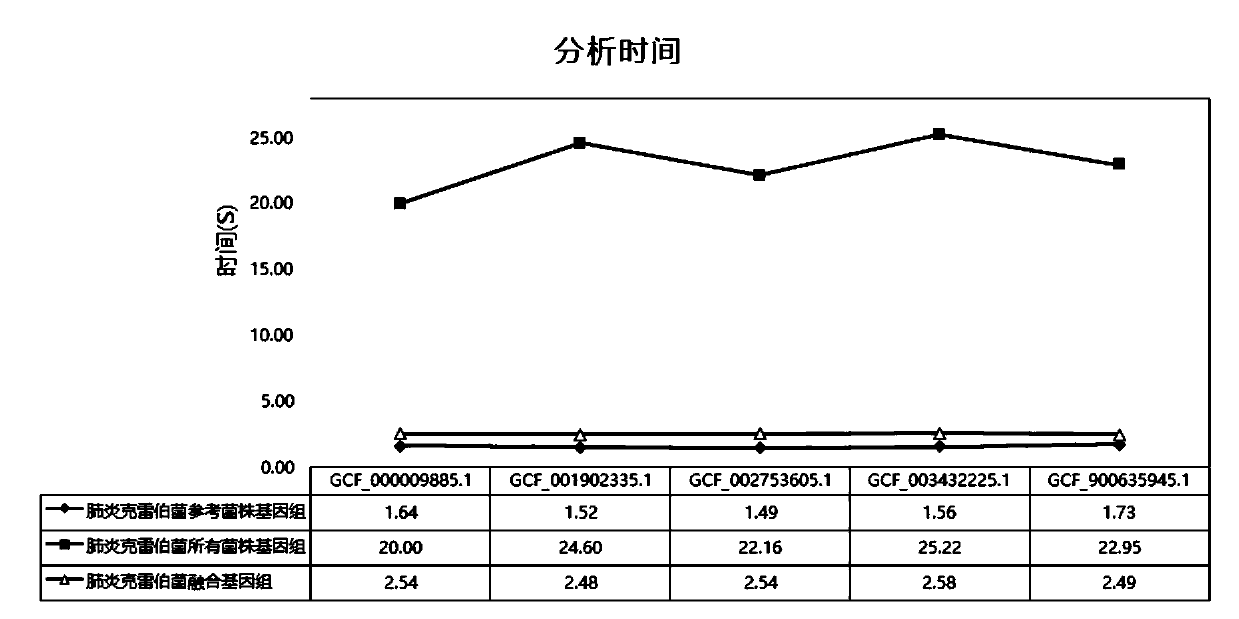
[0076] In order to evaluate the effect of the fusion genome of the Klebsiella pneumoniae constructed in the above-mentioned embodiment 1, all untreated strain genomes of Klebsiella pneumoniae, the NCBI reference strain genome of Klebsiella pneumoniae, the above-mentioned Klebsiella pneumoniae The accuracy and analysis time of the fusion genome of the bacteria were analyzed and compared.
[0077] 1. Data volume evaluation and comparison
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com