Construction and application of novel transgenic plant marker gene
A marker gene and transgene technology, applied in the field of plant biology, can solve the problems of weak gene translation initiation ability, large polycistronic expression structure, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Embodiment 1: marker gene Hygfpbar construction
[0017] Hygromycin resistance gene (HPT), green fluorescent protein gene EGFP and glufosinate resistance gene Bar were serially connected by overlap extension PCR. The designed primers are: HPTZ1: 5'-CCGCTCGAGATG AAAAAGCCTGAACTCACCG-3' (shown in SEQ ID NO.3); HPTF1:5'-ACATCAC CAGCAAGCTTAAGAAGATCGAAATTCAACAGCTATTTCTTTGCCCTCGG ACG-3' (shown in SEQ ID NO.4); GFPZ: 5' -CTTAAGCTTGCTGGTGATGTTGA ATCCAACCCAGGTCCAATGGTGAGCAAGGGCGAGGAG-3'(SEQ ID NO.5所示);GFPF:5'-ACATCACCAGCAAGCTTAAGAAGATCGAAATTCAACAGTCAGATCTCGGTGACGGGCAG-3'(SEQ ID NO.6所示);BarZ:5'-CTTAA GCTTGCTGGTGATGTTGAATCCAACCCAGGTCCAATGAGCCCAGAACG ACGCCCG-3'(SEQ ID NO .7); BarF: 5'-CCGCTCGAGTCAGATCTCGG TGACGGGCAG-3' (shown in SEQ ID NO.8).
[0018] In a 50 µl reaction system, the HPT gene was amplified from pCAMBIA1301 using primers HPTZ1 and HPTF1; the Bar gene was amplified from pPZp-RCS2-Bar (Novagen, Madison, USA) using primers BarZ and BarF; the Bar gene was amplified from ...
Embodiment 2
[0019] Embodiment 2: Construction of plant expression vector
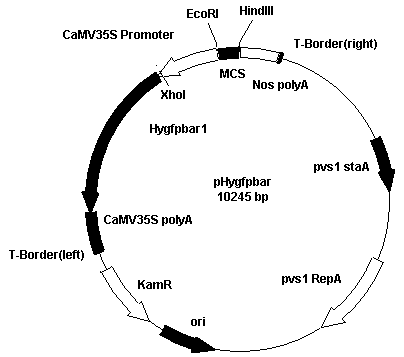
[0020] Firstly, the plant expression vector pCAMBIA1301 was double-digested with BglII and BstEII, and T4 DNA polymerase (Takara Baobiology) in the end smoothing kit was added, and then 5 U of T4 DNA ligase was added to identify the presence of the GUS gene in the vector by PCR, and screened to remove The vector of GUS gene is used as a novel marker gene expression vector. The pCAMBIA1301-GUS vector and the Hygfpbar fragment were digested with XhoI, and the recovered vector and fragment were ligated at a molar ratio of 1:10 or more. PCR was used to identify the direction of the inserted gene, and the clone of the hygromycin resistance gene on the promoter side was found for further identification, and a new marker gene plant expression vector pHygfpbar controlled by the CaMV35S promoter was constructed.
Embodiment 3
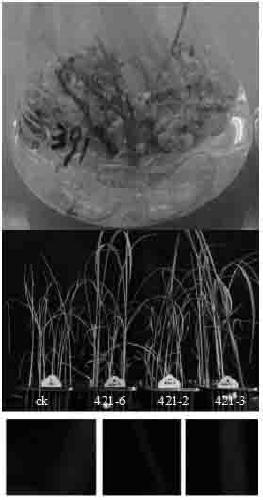
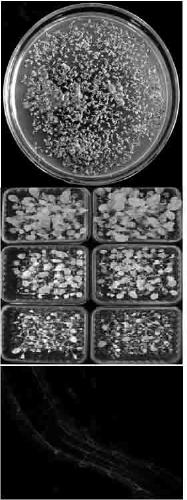
[0021] Example 3: Transformation of Rice and Arabidopsis
[0022] The strain used was Agrobacterium tumefaciens. The plasmid was introduced into Agrobacterium by electroporation. Pick a single bacterium into 25 ml YEB medium (50mg / l rifampicin) for overnight culture, transfer 5 ml of the bacterial liquid to 100 ml YEB medium (50mg / l rifampicin), and cultivate to OD 600 = 0.7-0.8, put the bacterial solution on ice for 10 minutes, centrifuge at 5000 rpm for 10 minutes, and collect the bacterial cells at 4°C, add 100ml sterile double distilled water to wash twice. Add 2 ml of 10% glycerol to suspend the bacteria and transfer to a 50 ml centrifuge tube. Centrifuge at 5500 rpm for 10 min at 4°C. Collect the cells, add 500 µl of 10% glycerol to suspend the cells, and transfer to a 1.5 ml centrifuge tube. Take 70µl competent cells and add 1µl recombinant plasmid pHygfpbar respectively. Mix well with a 200µl pipette tip removed, and transfer to a 0.1cm electric shock cup. Electri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com