Protein-ligand binding free energy calculating method based on MM/PBSA model
A computing method and protein technology, applied in the fields of proteomics, genomics, instruments, etc., can solve the problems of low computing efficiency, high-throughput computing without researchers, large computing errors, etc., to improve computing accuracy, eliminate deviation, the effect of improving computational efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The following will clearly and completely describe the technical solutions in the embodiments of the present invention with reference to the accompanying drawings in the embodiments of the present invention. Obviously, the described embodiments are only some, not all, embodiments of the present invention. Based on the embodiments of the present invention, all other embodiments obtained by persons of ordinary skill in the art without making creative efforts belong to the protection scope of the present invention.
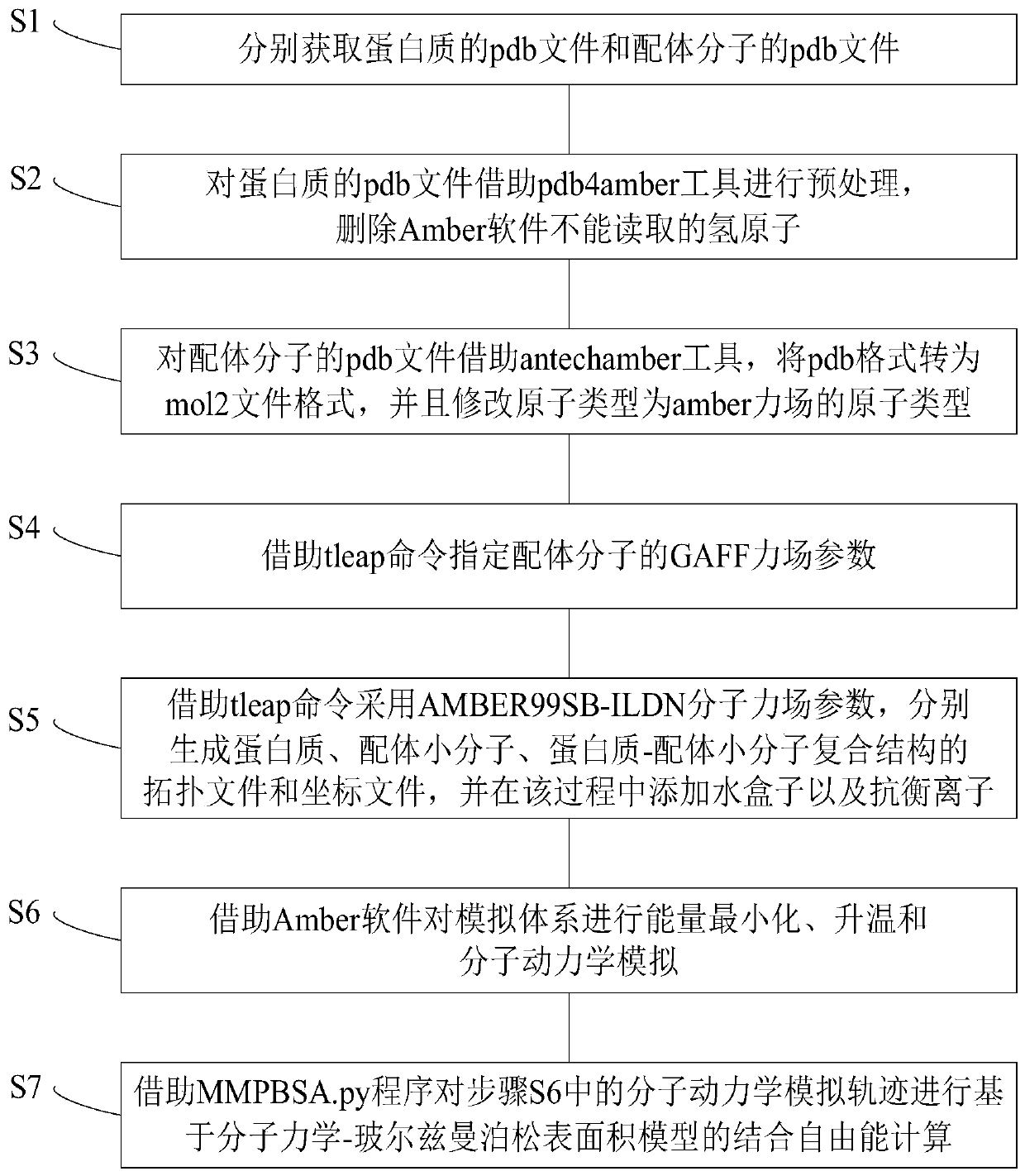
[0026] figure 1 It is a flow chart of the calculation method of protein-ligand binding free energy based on the MM / PBSA model of the embodiment of the present invention.
[0027] Such as figure 1 As shown, the protein-ligand binding free energy calculation method based on the MM / PBSA model of the embodiment of the present invention includes the following steps:
[0028] S1, obtain the pdb file of the protein and the pdb file of the ligand molecule respective...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com