Novel goose astrovirus SYBR Green dye method fluorescent quantitative PCR detection kit
A detection kit and fluorescence quantitative technology, applied in the field of poultry virus detection, can solve the problems of high technical requirements, high cost, unfavorable large-scale clinical sample detection, etc., and achieve the effects of improving detection efficiency, reducing losses, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Design and synthesis of primers
[0048] The genome sequences of multiple strains of NGAstV were selected from GenBank, including NGAstV-GD strain (accession number MG934571.1), NGAstV-SDPY strain (accession number MH052598.1), NGAstV-CHN / SD01 strain (accession number MF772821.1), NGAstV-HN1G strain (accession number KY807085.1), NGAstV-FLX strain (accession number NC_034567.1), NGAstV-CXZ strain (accession number MH807626.1) and NGAstV-AHDY strain (accession number MH410610.1), using Megalign The software compared and analyzed their conserved sequences, and found that the ORF1b gene sequences of each isolate were the most conserved overall. We selected some of the conserved regions for primer design. The ORF1b gene sequence was used as a template. In order to distinguish the NGAstV-SDPY strain from other goose astroviruses, we further selected a partial sequence in the ORF1b gene sequence of the NGAstV-SDPY strain (preservation number CCTCC NO: V201808) for ...
Embodiment 2
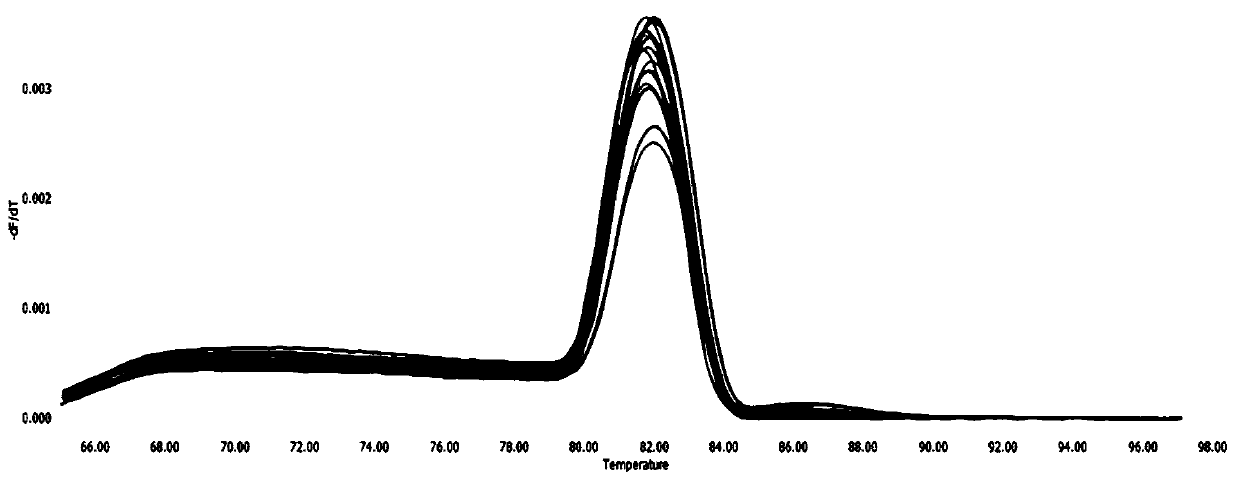
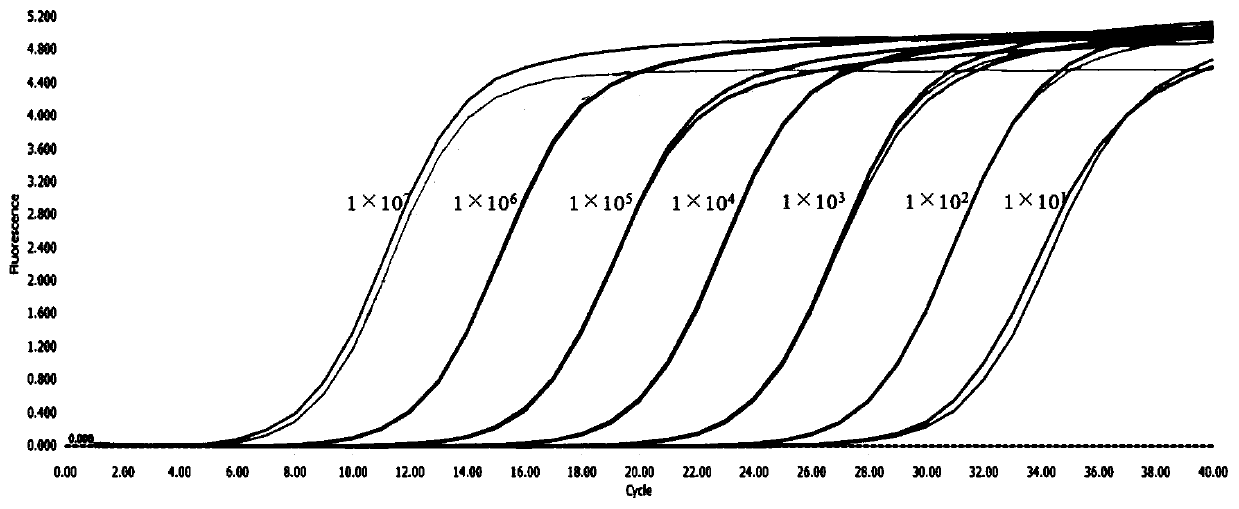
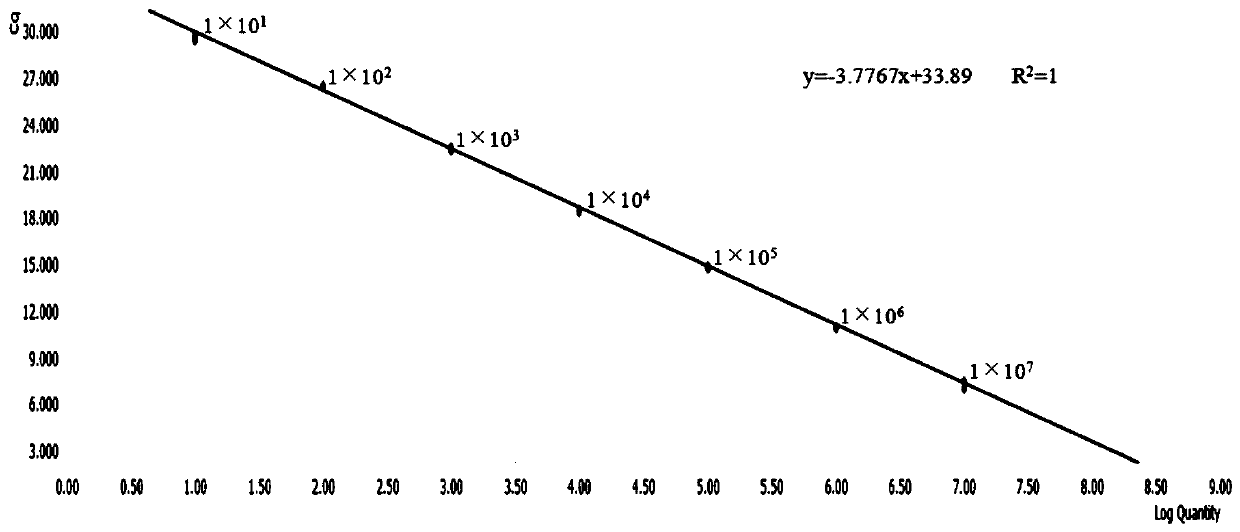
[0052] Example 2: The composition of the kit, the optimization of experimental parameters and the investigation of specificity, sensitivity and repeatability
[0053] 1. The composition of the kit:
[0054] The kit in this embodiment is a fluorescent quantitative PCR kit for detecting NGAstV. The kit contains: the upstream primer (10 μM) shown in SEQ ID NO.1 designed in Example 1, the downstream primer (10 μM) shown in SEQ ID NO.2 designed in Example 1, standard plasmid and fluorescent quantitative PCR Reagents.
[0055] Standard plasmids are prepared by the following methods:
[0056] The NGAstV genome is used as a template and the primers shown in SEQ ID NO.1-SEQ ID NO.2 are used to obtain an amplification product, the nucleotide sequence of which is shown in SEQ ID NO.3.
[0057] The amplified product of NGAstV was connected to the pMD18-T vector according to the operating procedures in the manual. The reaction system was: 4 μL of cDNA fragment, 1 μL of pMD18-T vector, a...
Embodiment 3
[0073] Embodiment 3: clinical application experiment
[0074] The diseased foie gras and kidney tissues with clinically suspected NGAstV infection were collected and homogenized, and the total RNA was extracted with TRIzol reagent. The RNA was reverse-transcribed into cDNA according to the operation steps using the kit, and stored at -20°C for later use.
[0075] Add 10 μL of TB Green fluorescent dye, 0.3 μL (10 μM) of upstream and downstream primers, 1 μL of cDNA template, and 8.4 μL of sterilized water into the fluorescent quantitative PCR tube, and the total reaction system is 20 μL. The reaction was carried out in a Roche LightCycle96 fluorescent quantitative PCR instrument, and the reaction conditions were: 95°C for 30s; 95°C for 5s; 60°C for 30s for 40 cycles. At the same time, standard products were added to make a standard curve.
[0076] Result Judgment Method: If the Ct value of the test sample is ≤30, and the amplification curve is a smooth "S" shape, it can be jud...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com