Method of quantifying mutant allele burden of target gene
An allele and target gene technology, applied in biochemical equipment and methods, microbial assay/inspection, etc., can solve problems such as underestimation of allele burden
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
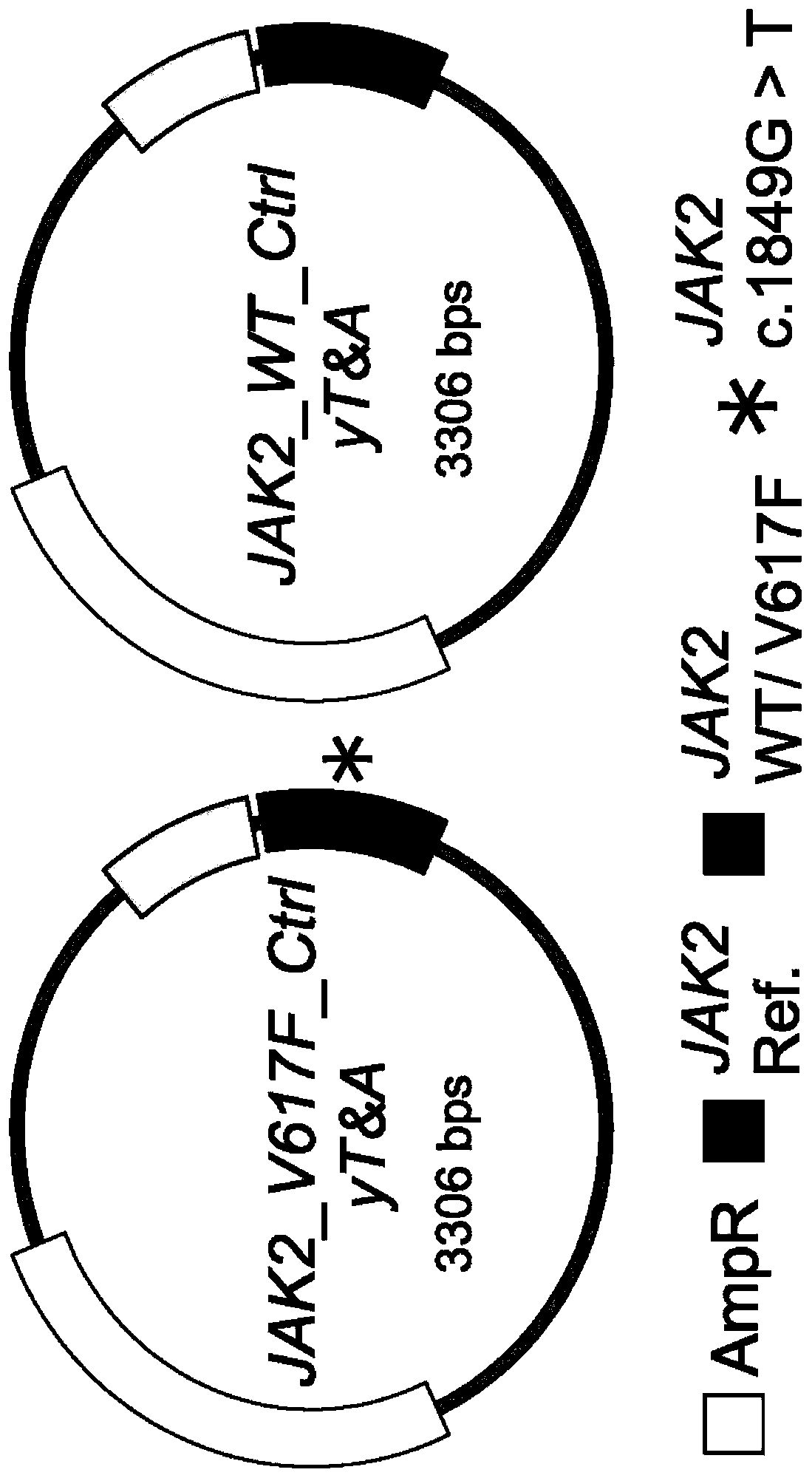
[0083] Example 1. Construction of a recombinant plasmid as a standard for quantifying JAK2 V617F mutant allele burden (mutant allele burden)
[0084] To minimize copy number variation and normalize the quantification of JAK2 V617F allele burden, the pair recombinant plasmids of the present invention were constructed as follows.
[0085] Since fewer somatic mutations are present in exon 21 of the JAK2 gene, applicants chose this region as an internal control. use TRI Genome DNA extracted from HEL cells was used as a template and PCR was carried out using the forward primer H_JAK2_clon_exon21_F and the reverse primer H_JAK2_clon_exon21_R shown in Table 1, and the control PCR product (264bp) with JAK2 exon 21 partial DNA sequence It was obtained. The resulting control PCR product was purified and then ligated into a cloning vector using the yT&A cloning kit (Yeastern Biotech, Taipei, Taiwan, China) according to the manufacturer's instructions to obtain the plasmid JAK2_Ctrl_yT...
Embodiment 2
[0090] Example 2. Verification of the accuracy of the recombinant plasmid of the present invention as a standard for quantifying JAK2 V617F mutant allele burden
[0091] experimental method:
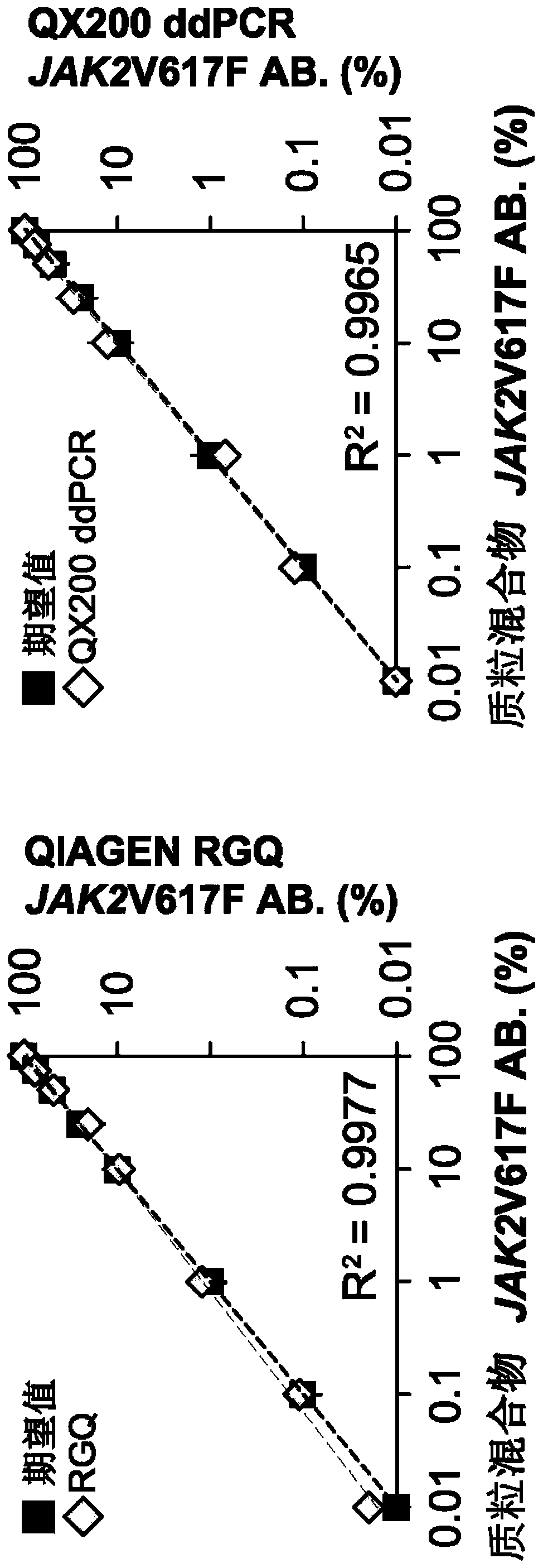
[0092] The wild-type and mutant target recombinant plasmids JAK2_WT_Ctrl_yT&A and JAK2_V617F_Ctrl_yT&A (representing 0% and 100% JAK2 V617F mutant allele burden, respectively) obtained from Example 1 were obtained in various different ways as shown in Table 2 below. ratio to obtain six recombinant plasmid mixtures (i.e., 100%, 50%, 10%, 1%, 0.1% and 0.01%) each with a different JAK2 V617F mutant allele burden, which Supplied as a standard dilution for the JAK2 V617F mutant allele.
[0093] Table 2
[0094] JAK2 V617F mutant allele burden (%) 100 50 10 1 0.1 0.01 JAK2_WT_Ctrl_yT&A (μL) 0 50 90 99 99.9 99.99 JAK2_V617F_Ctrl_yT&A (μL) 100 50 10 1 0.1 0.01
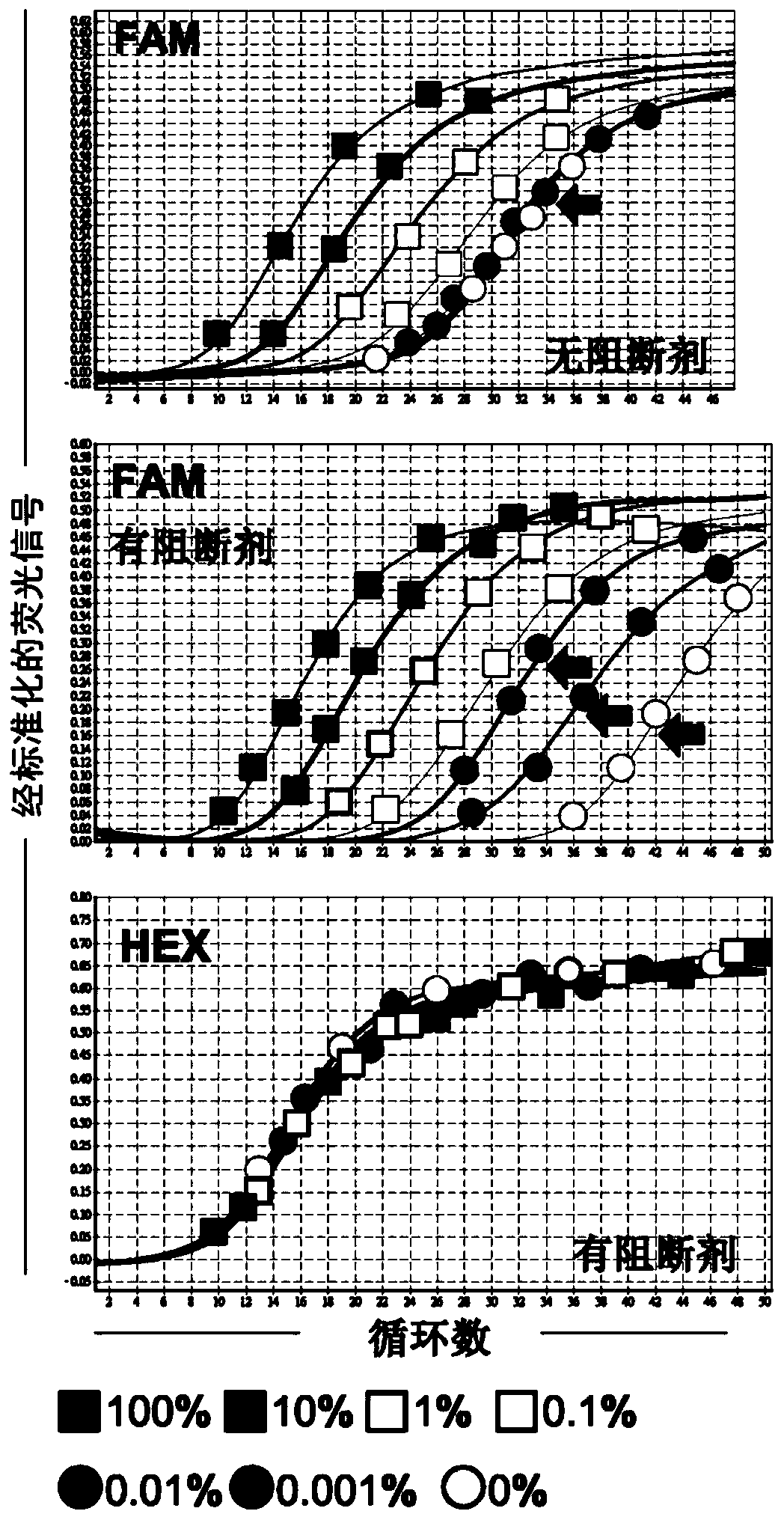
[0095] The JAK2 V617F mutation in the above-mentioned recombinant plasmid mixture was test...
Embodiment 3
[0099] Example 3. Quantification of JAK2 V617F mutant allele burden in recombinant plasmid mixtures and clinical samples by quantitative duplex PCR assay
[0100] A. Establishment of standard curve for JAK2 V617F mutant allele burden
[0101] 1. Plasmid-based standard curve:
[0102] Seven recombinant plasmid mixtures (with JAK2 V617F mutant allele burdens of 100%, 10%, 1%, 0.1%, 0.01%, 0.001% and 0%, respectively) were prepared according to the method described in Example 2. is prepared. Each of these recombinant plasmid mixtures was used as a DNA template (template) for quantitative double-PCR analysis, which was performed in Rotor-Gene Q (RGQ) 5plexHRM using the PCR reaction mixture (PCR reaction mixture) and reaction conditions shown in Table 3. Platform (Rotor-Gene Q 5plex HRM platform) (Cat No.: 9001580, Qiagen, Germany) was carried out. JAK2_exon 14 mutant allele-specific primer pairs and probes and JAK2_exon21-specific primer pairs and probes as listed in Table 4 we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com