Method and device for detecting chromosomal aberrations
A detection method and chromosome technology, applied in the field of chromosome detection, can solve problems such as data correction errors, sex chromosome abnormalities and chimera detection effects are not obvious, and chromosome abnormality signals are missed, so as to reduce false positives and false negatives, reduce Probability of missed detection of chromosomal abnormalities and the effect of high detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
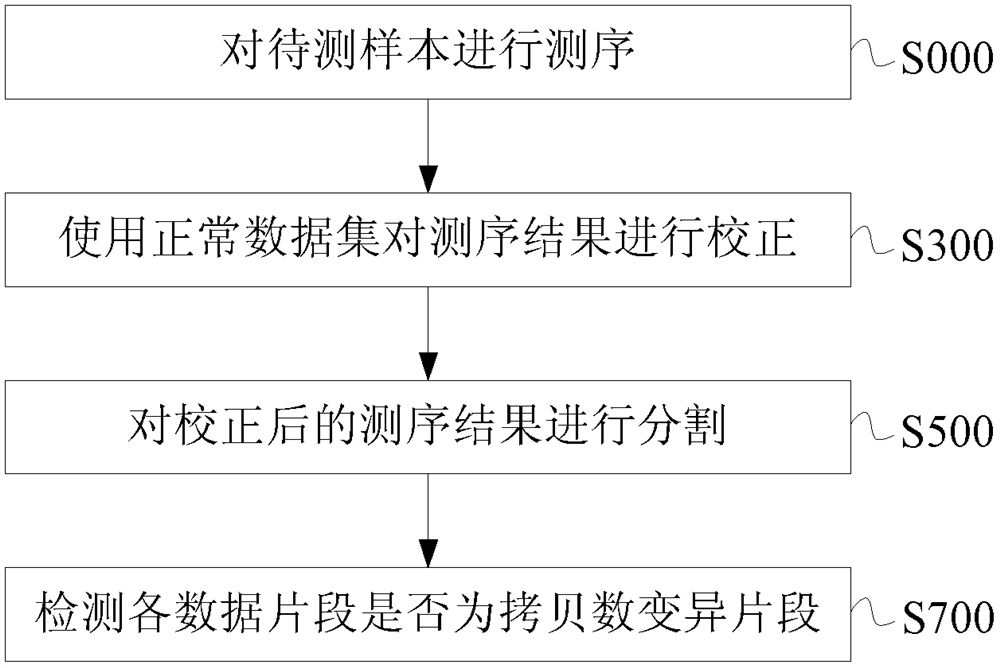
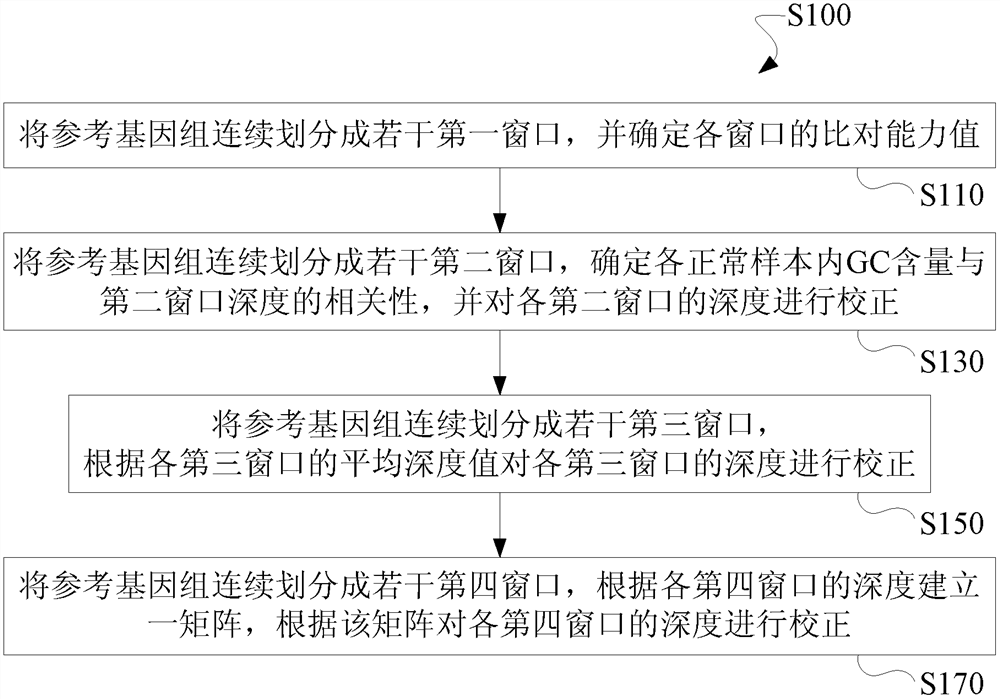
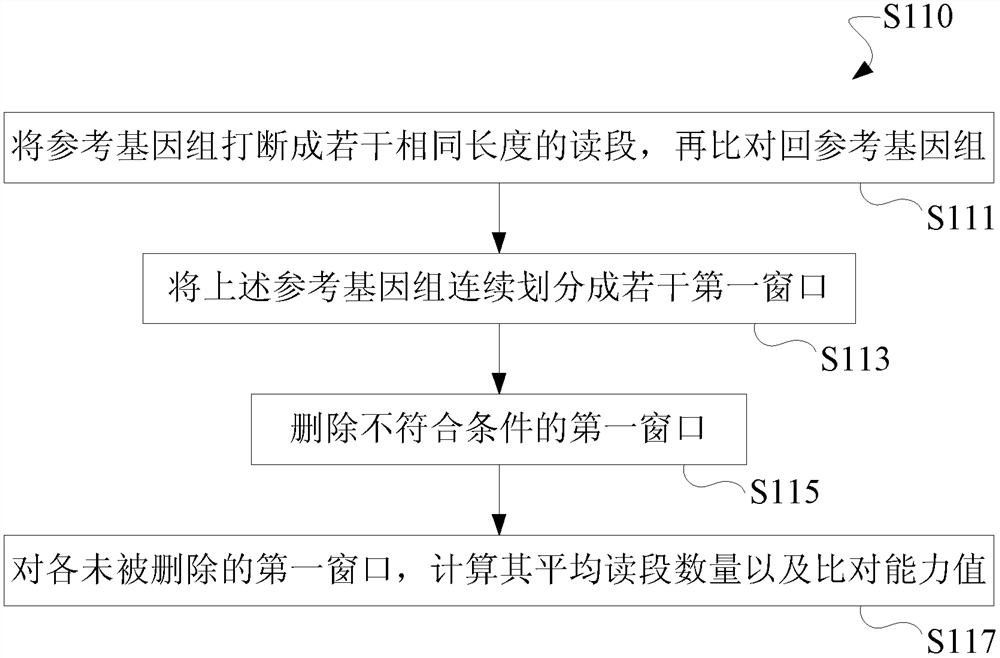
[0149] In view of the problems existing in the prior art solutions, the present invention overcomes the defects in the existing data correction methods, reduces the probability of missing detection of chromosomal abnormalities due to the use of the same batch of samples for comparison in the prior art; False-positive and false-negative results of test results caused by the bias between Chimera detection; improved detection of chromosomal copy number variations below 10Mb and at low free nucleic acid ratios; reduced data bias and the resulting false positive and false negative rates of test results.
[0150] the term
[0151] Sample: The sample described in the present invention is a biological sample containing nucleic acid.
[0152] Normal sample: The normal sample in the present invention is a sample whose karyotype is found to be normal by amniocentesis or chorionic villus sampling, and it is determined by the prior art that it does not have chromosome number variation and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com