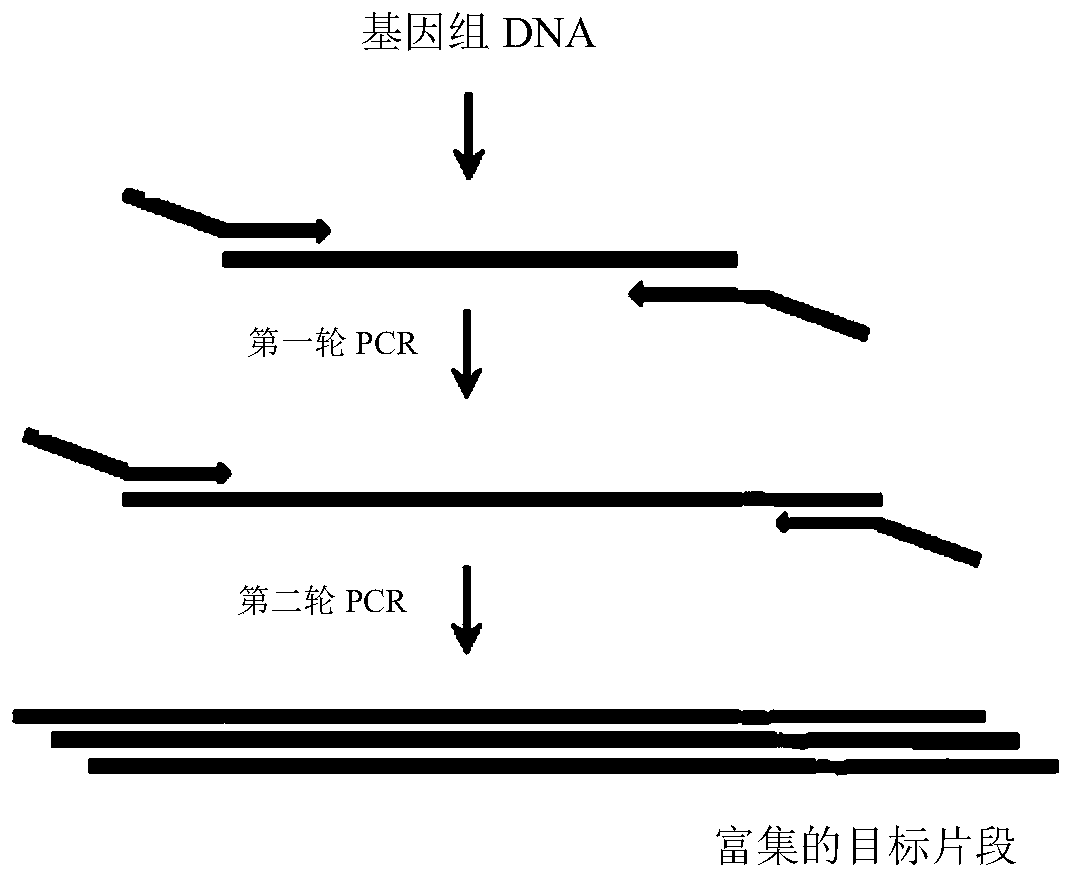
Method for enriching target DNA fragments by multiplex PCR
A technology of target fragments and fragments, applied in DNA/RNA fragments, recombinant DNA technology, DNA preparation, etc., can solve problems such as high cost, large amount of starting genomic DNA, and reduced sequencing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
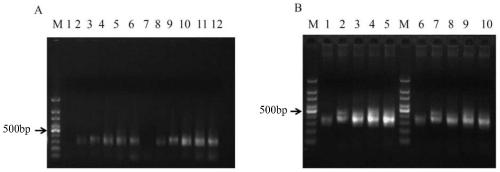
[0111] Embodiment 1, utilize multiplex PCR to enrich the target segment of rice genomic DNA
[0112] 1.1 Experimental materials
[0113] Genomic DNA of Zhonghua 11 (ZH11). Genomic DNA was extracted using the standard CTAB method, and the final concentration was diluted and quantified to 50ng / μL.
[0114] 1.2 Preparation of multiplex PCR primers
[0115] Two rounds of PCR primers were designed and each primer was synthesized.
[0116] Design of the first round of PCR primers (1st PCR): primers were designed for 24 known SNP sites of rice, and the size of the genome amplified by the primers of each site was 200-250bp. The 5' end of the forward primer at each site all contains a common sequence of 21bp (this sequence is an overlapping sequence (overlap) in the Illumina sequencing primer, TACACGACGCTCTTCCGATCT), and the rest is used to identify rice genomic DNA; The 5' ends all contain a 21bp common sequence (the sequence is an overlapping sequence (overlap) in the Illumina se...
Embodiment 2
[0181]Example 2, Optimization of the method for multiple PCR enrichment of target fragments
[0182] It can be concluded from the results of Example 1 that the more the number of primers increases, the worse the effect of enriched target fragments is, especially the homogeneity among the enriched target fragments is very poor. In order to solve the problem of the uniformity of the target fragment obtained by enrichment, the inventors tested the effect of using 5 DNA polymerases and 6 primer pairs to amplify the target fragment by multiplex PCR.
[0183] 2.1 Experimental materials
[0184] Genomic DNA of Zhonghua 11 (ZH11). Genomic DNA was extracted using the standard CTAB method, and the final concentration was diluted and quantified to 50ng / μL.
[0185] 2.2 Preparation of multiplex PCR primers
[0186] In order to reduce the amount of primers, three rounds of PCR amplification were used in this embodiment, three rounds of PCR primers were designed, and each primer was synt...
Embodiment 3
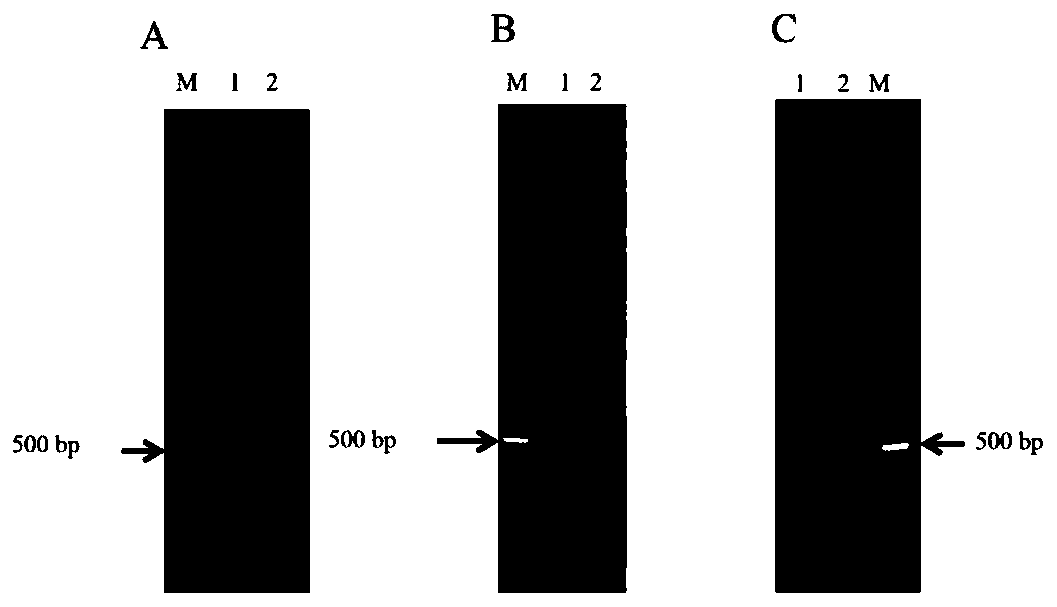
[0272] Embodiment 3, optimization of multiplex PCR
[0273] 1. Analysis of Factors Affecting Multiplex PCR
[0274] In order to achieve specific enrichment of more target fragments in a PCR reaction system, the inventors used different DNA sequence analysis software (DNAMAN software and Oligo7 software) to analyze the characteristics of the primers, specifically for the 3' end of each primer The number of 3 nucleotides or 4 nucleotides complementary to the 5' end and other primers in the same PCR reaction system was counted, and the complementarity between the primers was counted again using the PCR Primer Stats and Multiple Primer Analyzer websites , the analysis objects are 57 pairs of primer sequences composed of Table 2.1 and Table 2.2 in Example 2, and the indicators analyzed by each software and website are specifically shown in Table 3.1.
[0275] Using DNAMAN software and Oligo7 software to count the factors affecting PCR amplification of each pair of primers (Table 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com