Blocking type PD-L1 camel-source single-domain antibody and use thereof
A PD-L1, single-domain antibody technology, applied in the field of biomedicine or biopharmaceuticals, can solve problems such as the reduction of T cell immune effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0139] Example 1: Construction, screening and identification of PD-L1 single domain antibody library
[0140] Library construction:
[0141] (1) Mix 1 mg of hPD-L1(ECD)-Fc antigen with Freund's adjuvant in equal volume, immunize a Xinjiang Bactrian camel once a week, and immunize 7 times in total to stimulate B cells to express antigen-specific single domain Antibody; (2) After 7 times of immunization, extract 100mL camel peripheral blood lymphocytes and extract total RNA; (3) Synthesize cDNA and amplify VHH by nested PCR; (4) Use restriction enzymes PstI and NotI Cut 20 μg of pMECS phage display vector and 10 μg of VHH and ligate the two fragments; (5) Transform the ligation product into electroporation-competent cells TG1 to construct a PD-L1 single domain antibody library.
[0142] Antibody Screening:
[0143] (1) 10 μg ghPD-L1(ECD)-Fc antigen (10 μg Fc inNaHCO 3 As a control) coupled to NUNC microtiter plates, placed overnight at 4°C; (2) the next day, 100 μL of 0.1% BS...
Embodiment 2
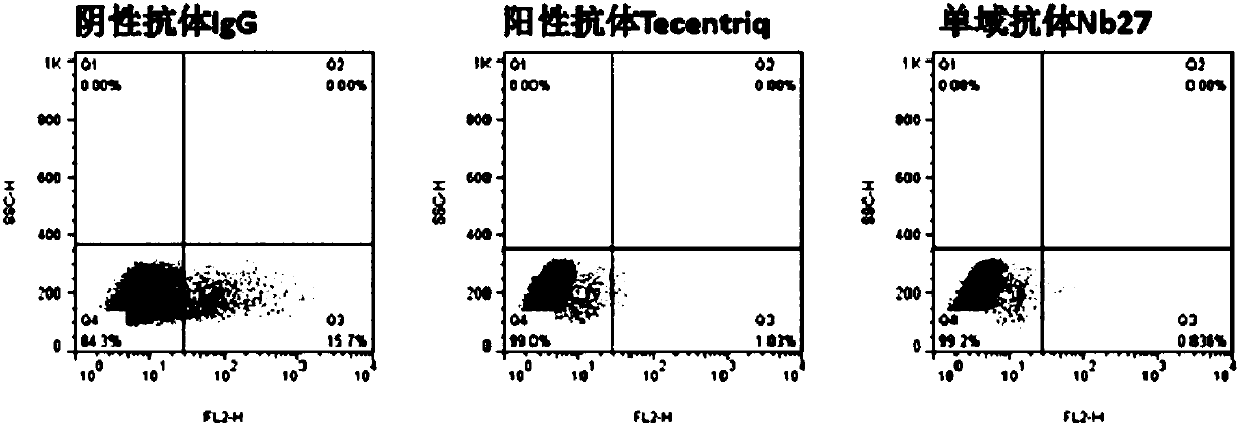
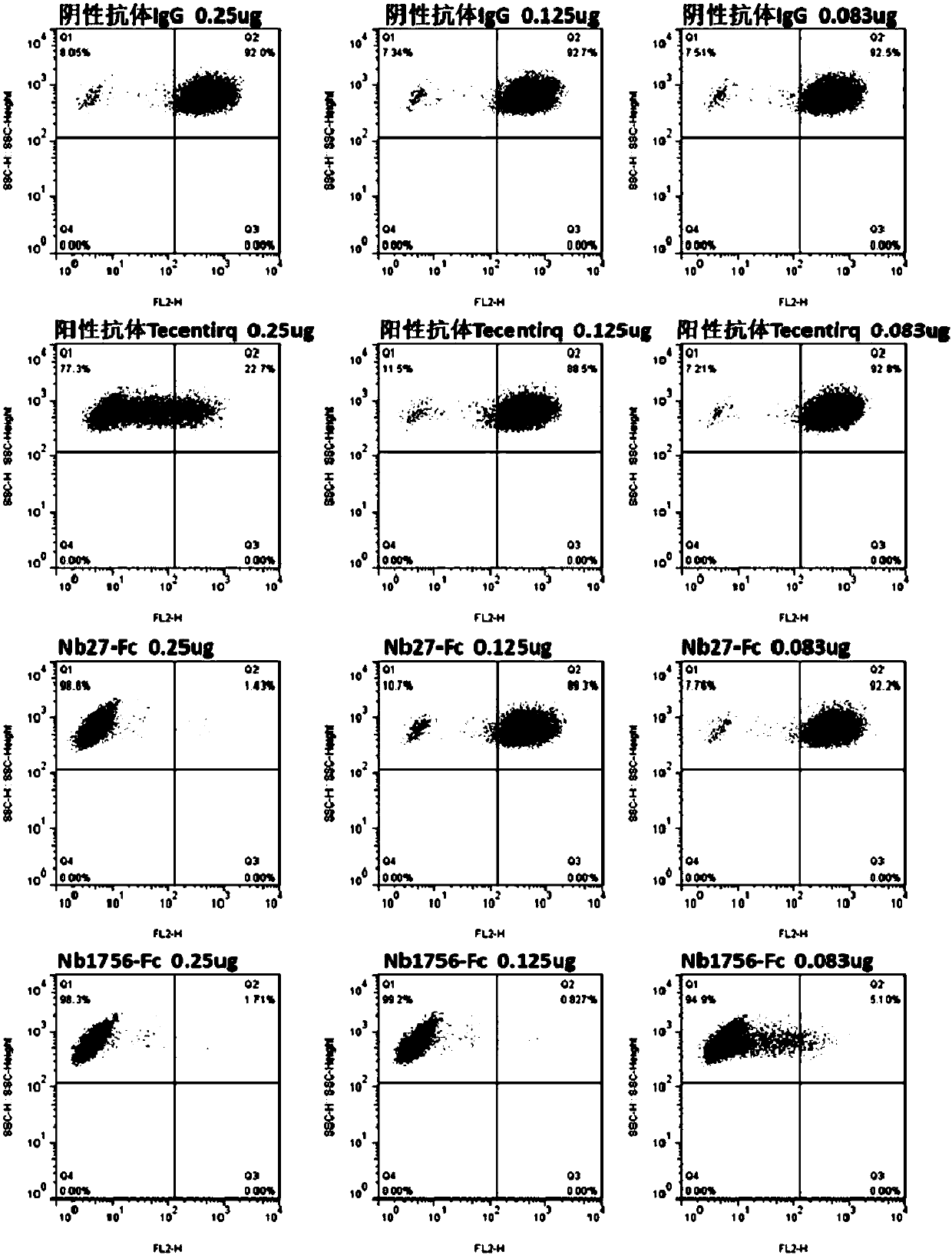
[0146] Example 2: Preliminary identification of blocking function of single domain antibody by flow cytometry
[0147] (1) Preparation of hPD-1(ECD)-Biotin protein (the preparation method of hPD-1(ECD) is the same as in Example 1), and the method of protein biotinylation refers to the instructions of biotin reagent; (2) Transient PD-L1 gene Express PD-L1 protein on the cell surface of HEK293F cells; (3) Prepare crude lysate of PD-L1 single domain antibody TG1 strain, the preparation method is the same as that of Zhu Min et al., Nanoscale Res Lett., 2014Sep26; 9(1) : 528; (4) Each sample takes 1×10 6 HEK293F cells transiently transfected with PD-L1 were resuspended in 0.5% BSA-PB Sbuffer, 100 μL of the above crude extract was added, and negative control (hIgG1) and positive control (Tecentriq) were set at the same time, and 5 μghPD-1 (ECD) was added to each well -Fc-Biotin, incubated at 4°C for 20 min; (5) Wash the cells twice with PBS, add eBioscience SA-PE, incubate at 4°C f...
Embodiment 3
[0148] Example 3: Humanized transformation of PD-L1 blocking single domain antibody
[0149] (1) First use the PD-L1 single domain antibody sequence shown in SEQ ID NO.:8 as a template to search for homologous structures in the structural database, and take the structure where Evalue=0.0 and sequence identity ≥ 70%; (2 ) Structural comparison of these structures, and according to the crystal structure resolution size and the constructed phylogenetic tree, multi-template homology modeling based on the PD-L1 single domain antibody sequence shown in SEQ ID NO.:8, and then According to the ranking of the scoring function, select the structure with the lowest molpdf; (3) For the optimal structure modeled, use the ProtSA server to calculate the solvent accessibility of the residue, that is, the solvent accessibility of the folded state of the residue relative to the unfolded state. The ratio of the contact area is the criterion, and the residues greater than 40% are taken as the res...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com