Method for initiating rolling circle amplification and FRET to detect miRNA based on Toehold-mediated strand displacement reaction
A chain displacement reaction and detection method technology, applied in the field of molecular detection, can solve the problem of low signal output and achieve good selectivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
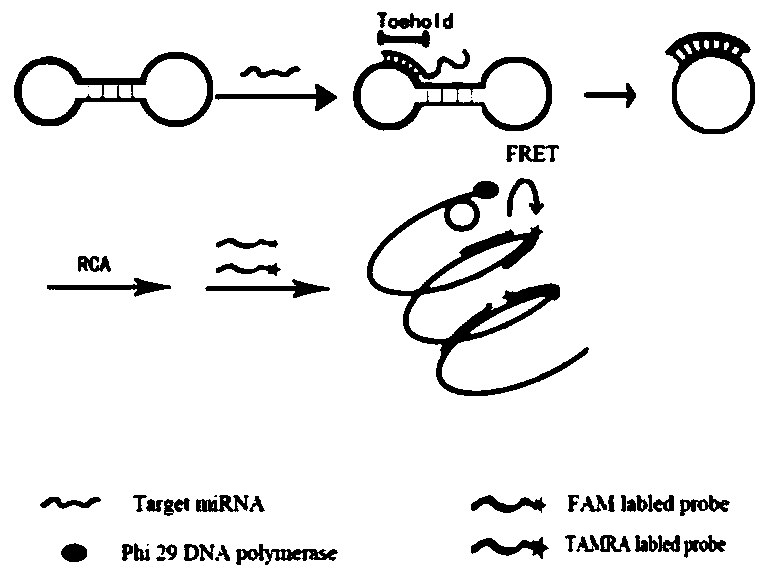
[0052] The principle of this embodiment is as figure 1 As shown, the experimental procedure is as follows.
[0053] Preparation of dumbbell-shaped DNA probes
[0054] Prepare 20 μL reaction system for each DNA probe of T6, T7, T8, T9 and T10, including 1 μL DNA probe (100 μM), 2 μL 10×T4 DNA ligase reaction buffer (500 mM Tris-HCl, 100 mM MgCl 2 , 100 mMDTT, 10 mM ATP, pH 7.5, 25°C), 1 μL T4 DNA ligase (400 U / μL), 16 μL DEPC water. The system was reacted at 23°C for 2 hours, and then heated at 65°C for 10 minutes to terminate the reaction. Add 3 μL NEB buffer Ⅰ (100mM BisTris Propane-HCl, 100mM MgCl 2 , 10mM DTT, pH 7.5, 25℃), 1μL exonuclease Ⅰ (20U / μL), 1μL exonuclease Ⅲ (100U / μL), incubate at 37℃ for 30 minutes, then heat at 80℃ for 20 minutes Enzyme inactivation. The prepared dumbbell-shaped DNA probes were stored at -20°C.
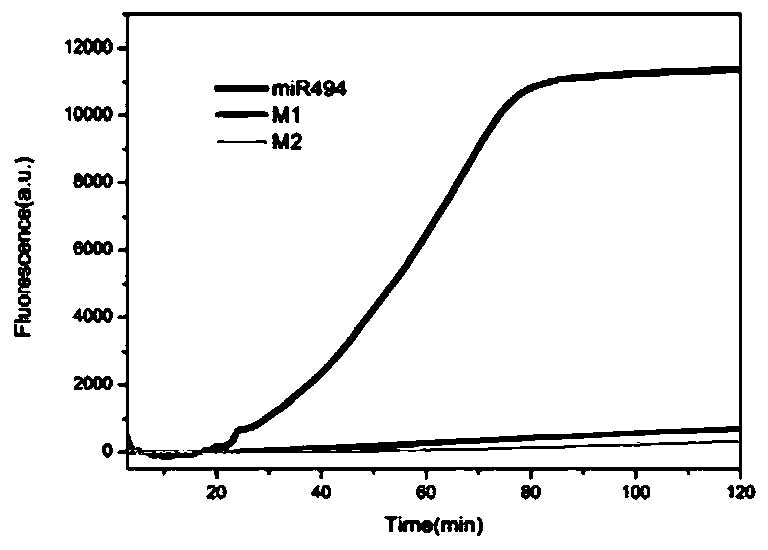
[0055] Real-time fluorescence detection of dumbbell-shaped DNA probe using TMSD reaction to initiate RCA reaction (hereinafter referred to as TI...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com