Detecting method for accurately and rapidly detecting target superbacteria
A super bacteria, detection target technology, applied in the medical and health field, can solve the problems of hidden dangers to the health of patients and medical staff, difficult to completely remove, etc., to achieve rapid and accurate detection and judgment, good application value, and reduce the effect of operation difficulty.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
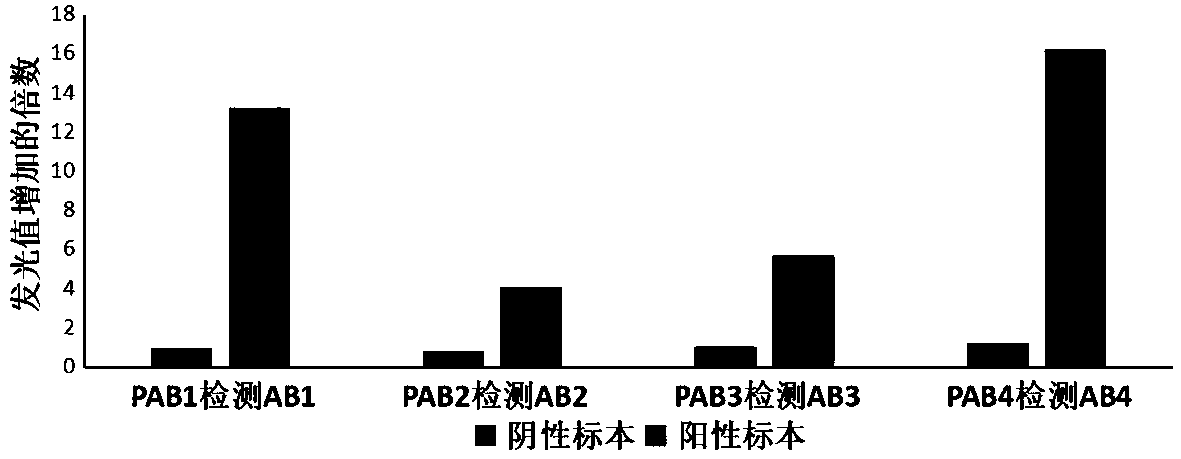
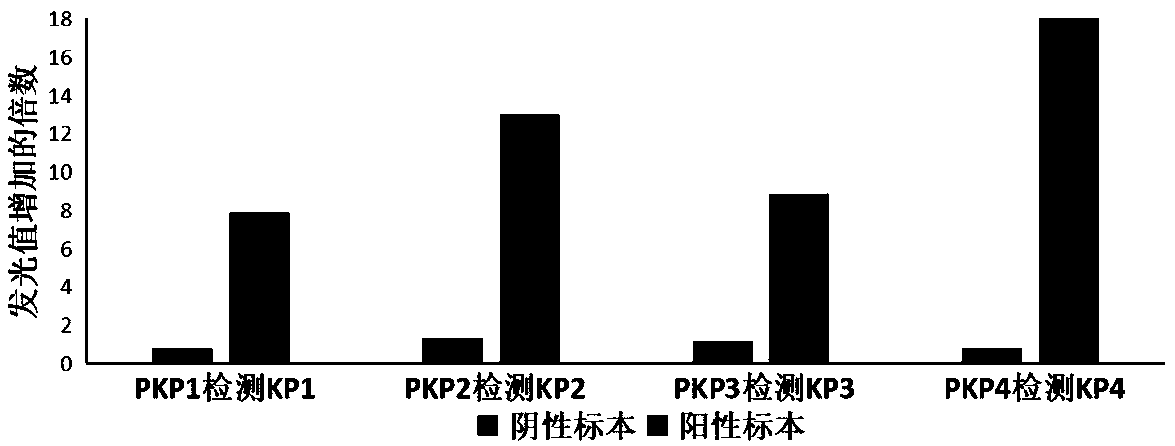
[0052] In this embodiment, the qualitative detection and determination of super bacteria using fluorescence luminescence characteristics is used as an example, and the relevant detection process is briefly introduced as follows.
[0053] It should be noted that, in order to further prove the specificity of phages, the samples prepared during the experiment were mixed samples (that is, the mixed samples were a mixture of multiple super bacteria).
[0054] (1) Mixed samples and enriched culture
[0055] The common superbug standard samples (screened, identified and stored in advance) were inoculated into sterile LB medium (containing 16ug / ml of carbapenem antibiotic imipenem), and the mixed culture was used as a mixed sample.
[0056] In this example, during mixed culture, the general culture conditions are: 37°C, 200rpm shaking culture until the super bacteria concentration is not less than 10 6 CFU / ml;
[0057] The common target super bacteria, that is, the super bacteria i...
Embodiment 2
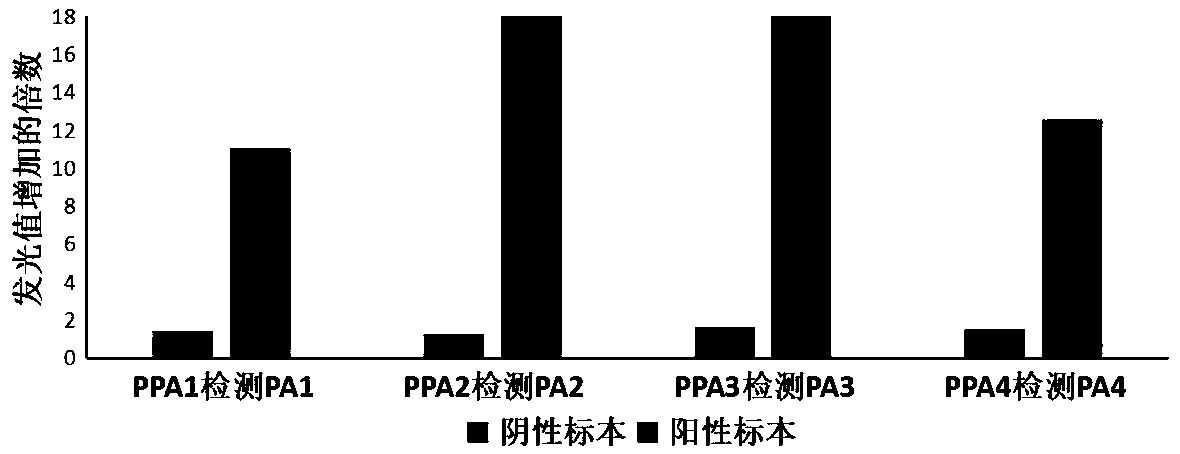
[0070] In this example, the construction of a semi-quantitative detection method for a specific superbacterium pan-drug resistant Acinetobacter baumannii AB09V (corresponding to phage ZZ1) is taken as an example, and the relevant detection process is briefly introduced as follows.
[0071] (1) Sampling and preparing mock samples
[0072] Use a planktonic air sampler to collect planktonic bacteria in the environment as a background sample (the background sample does not contain super bacteria), and add different concentrations in the early stage (the addition amount is 10 1 , 10 2 , 10 3 , 10 4 , 10 5 , 10 6 CFU / ml) pan-drug resistant Acinetobacter baumannii AB09V was prepared as a mock sample containing super bacteria (the sampling time reflects the added concentration of Acinetobacter baumannii AB09V), and preliminarily enriched in sterile LB medium , 37°C, 200rpm shaking culture, and sampling every 1h in the subsequent experiments to investigate the influence of differe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com