Set of HDACs nucleic acid aptamers
A nucleic acid aptamer and aptamer technology, applied in the fields of genetic engineering, immunology and oncology, to achieve the effect of small molecular weight, easy synthesis and modification, and easy synthesis and preservation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
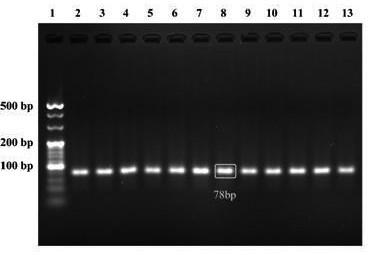
[0041] The HDACs nucleic acid aptamer of the present invention takes HDAC1 as an example, and the specific technical scheme is:
[0042] 1. Optimize the annealing temperature in the PCR program
[0043] The annealing temperature in the PCR program of the ssDNA library has a greater impact on the entire screening process. Therefore, in order to achieve the highest efficiency of PCR amplification in each round of screening, the annealing temperature in the PCR program of the ssDNA library needs to be optimized before the start of the screening. The specific PCR system is shown in Table 1:
[0044] Table 1 The optimized reaction system of PCR annealing temperature
[0045] Reaction component
volume
10x PCR Buffer
2 μL
dNTP (2.5 mM)
1.6 μL
FP (10 μM)
0.8 μL
RP (10 μM)
0.8 μL
Taq DNA Polymerase (5 u / μL)
0.2 μL
Template
2 μL
H 2 O
12.6 μL
[0046] The specific steps are as follows:
[0047] Prepare 12 tubes of PCR reaction mixture according to the table, mix well, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com