Bombyx mori microparticle hypothetical protein nb29 and its recombinant expression vector and application
A technology of silkworm microparticles and hypothetical proteins, which can be used in vectors, applications, nucleic acid vectors, etc., can solve the problems of inability to carry out virus-carrying analysis of progeny, high technical requirements for operators, and inability to accurately determine virus-carrying individuals.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1. Bioinformatics analysis and secretion verification of silkworm microparticles NB29
[0038] The nucleotide sequence of the silkworm microparticle hypothetical protein gene NB29 was obtained, and its sequence is shown in SEQ ID No.1. Use the corresponding website to predict nuclear localization signals, signal peptides and transmembrane domain analysis.
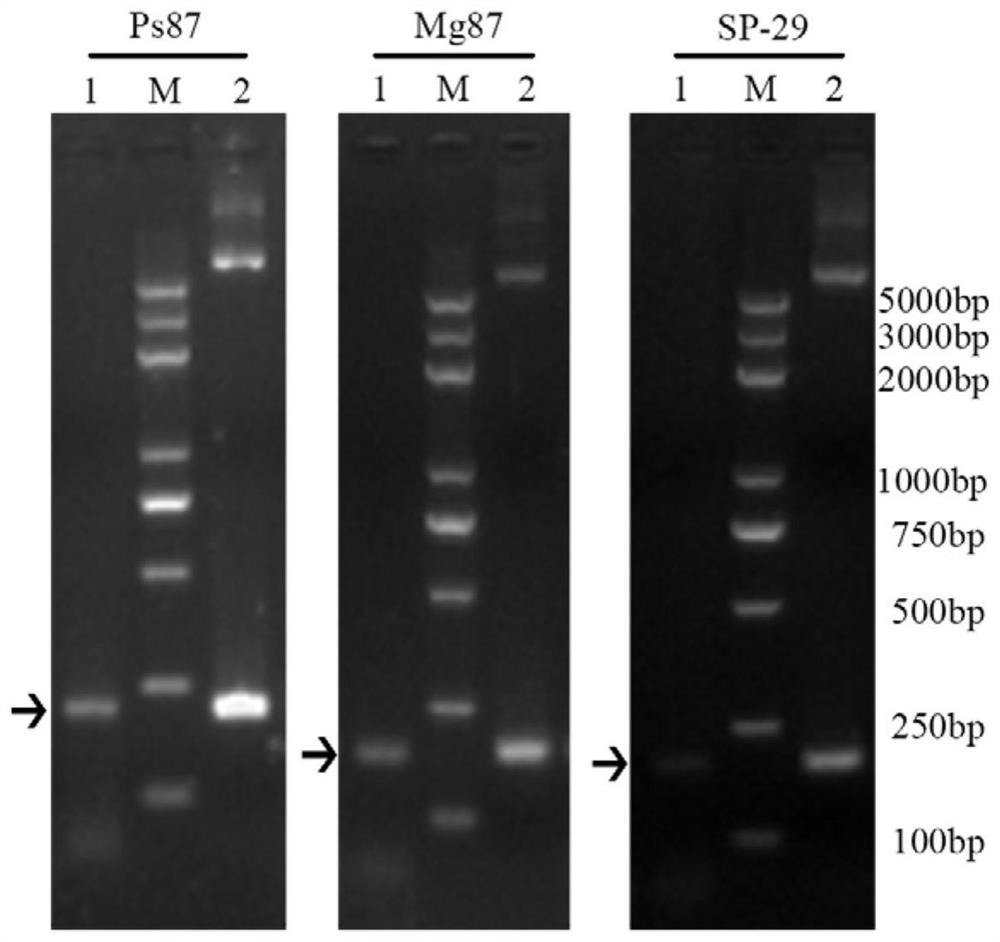
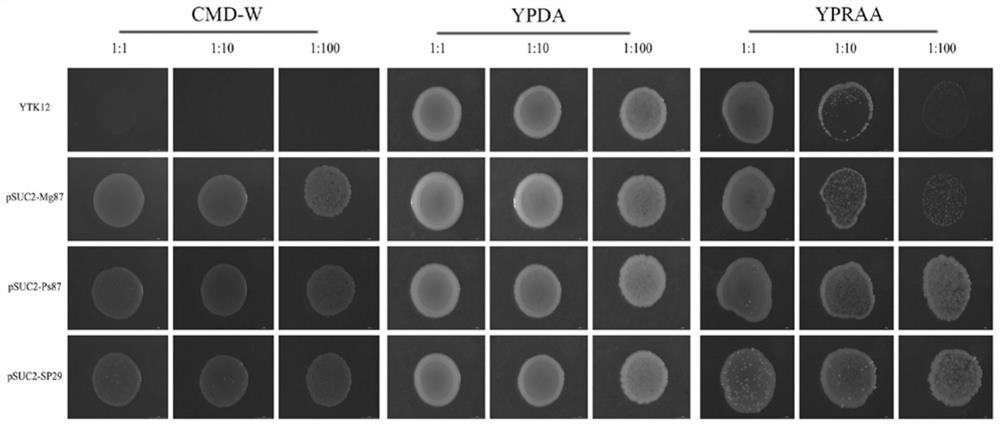
[0039] According to bioinformatics analysis, NB29 contains a signal peptide sequence with two nuclear localization signals, such as figure 1 shown. NB29, Ps87, and Mg87 signal peptide sequences were designed, the sequences of which are shown in SEQ ID No.2, SEQ ID No.3, and SEQ ID No.4, respectively, and then sent to Gensript to synthesize the sequences.
[0040]The synthetic signal peptide sequence was cleaved by Xba I and Kpn I restriction sites at the same time, and then ligated to the yeast signal sequence trap vector pSUC2T7M13ORI that had undergone the same digestion, and the ligated product was trans...
Embodiment 2
[0042] Example 2, identification of secreted protein NB29 self-interaction
[0043] From Example 1, it was verified that the silkworm microparticle NB29 is a secreted protein, which meets the requirement of being secreted into host cells. In order to further construct the system, NB29 is first truncated according to the structural domain, and the truncated NB29-A (full length), NB29-B, NB29-C, and NB29-D sequences are designed, and the sequences are as SEQ ID No.1, SEQ ID No.5, SEQ ID No.6, and SEQ ID No.7 are shown, and then sent to Gensript Company to synthesize the sequence. NB29-A, NB29-B, NB29-C and NB29-D utilize Kpn I and BamH I to connect to the pIZ-DsRed carrier (the DsRed fluorescent protein sequence as shown in SEQ ID No.9 is connected to Hind III and Kpn of the pIZ carrier 1 restriction site), the ligation product was transformed into Escherichia coli DH5α competent cells, then positive clones were screened with an LB plate containing bleomycin (Zeocin), a single ...
Embodiment 3
[0047] Example 3, Screening of secreted protein NB29 optimal truncation
[0048] On the basis of the self-interaction of the secreted protein NB29-B, the truncated NB29-N3 (SEQ ID NO.8) was further screened out and sent to Gensript to synthesize the sequence, in order to lay the foundation for the fusion of Cas9. First, pIZ-NB29-N3 was transfected into BmN-SWU1 cells, the method was similar to Example 2, the difference was that after blocking with the blocking solution, the cells were lightly washed with 1×PBST for 3 times, each time for 5 min; the mouse Myc antibody was incubated at room temperature for 1 h, and then washed with Wash cells lightly with 1×PBST 3 times, 5 min each time; add Alexa555-labeled goat anti-mouse secondary antibody IgG and nuclear dye DAPI, treat at 37°C in the dark for 1 hour, wash cells 3 times with 1×PBST lightly, 5 min each time . Take out the cover slip and observe its location in the cells by fluorescence, the results are as follows: Figure 8...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com