SYBR GreenI fluorogenic quantitative PCR detection method for hepatitis B virus
A hepatitis B virus, fluorescence quantitative technology, applied in the biological field, can solve the problems of high cost and unfavorable clinical specimen development
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] [Example 1] Target selection, primer design and screening
[0025] (1) Target selection and primer design:
[0026] Using the http: / / www.ncbi.nlm.nih.gov / blast website to search for the HBV C gene sequence from China, and using DNAMAN software for sequence comparison, find out the conserved region within the C gene range, design primers, Primer sequences are shown below;
[0027] Table 1 HBV primer pair and amplification length
[0028]
[0029] (2) Primer screening
[0030] Using pBS-HBV-3.6 Ⅱ plasmid as a template, PCR amplification was carried out with different primer pairs, and the amplification efficiency of the amplified products was detected by agarose gel electrophoresis. Screen out the best primer pair for subsequent experiments;
[0031] The PCR amplification system is 20 μL, and the specific components are: LA Taq MIX 10 μL, upstream primer 1 μL, downstream primer 1 μL, template 1 μL, nuclease-free water 7 μL;
[0032] The reaction program of PCR is:...
Embodiment 2
[0033] [Example 2] Hepatitis B virus SYBR Green Ⅰ fluorescent quantitative PCR detection method standard curve preparation
[0034] (1) Production of standard products
[0035] Use pBS-HBV-3.6 Ⅱ plasmid to make HBV detection standard, use NanoDrop Lite spectrophotometer to calculate the plasmid concentration, and calculate the copy number, the copy number calculation formula is: (6.02×10 23 )×(g / ml) / (DNA length×660)=copies / ml. Dilute the positive control plasmid in a 10-fold gradient to 10 3 ~10 8 Copies / ml 6 dilution gradients to make a standard curve;
[0036] (2) Standard curve creation
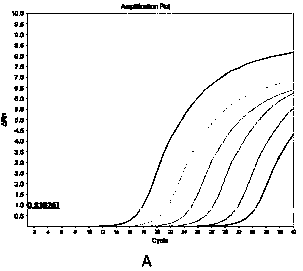
[0037] Take 1×10 3 ~ 1×10 8 Copies / mL 6 gradient standard products were amplified by fluorescent quantitative PCR to obtain the amplification curve, and the standard curve with the logarithm of the copy number of the initial concentration as the x-axis and the Ct value as the y-axis. The standard curve equation is y = 3.3521x +14.563, correlation coefficient R² = 0.9985.
Embodiment 3
[0038] [Example 3] Reproducibility verification of hepatitis B virus SYBR Green Ⅰ fluorescent quantitative PCR detection method
[0039] Selected recombinant plasmid standard products were divided into 1 × 10 5 ~ 1×10 7 3 concentrations of copies / mL are used to evaluate the repeatability of SYBRGreen Ⅰ fluorescence quantitative PCR. The Ct value among 3 parallel experimental tubes for each concentration. The results showed that the Ct values of the three parallel tubes in the same experiment basically coincided, indicating that the system had good repeatability.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com