Oligonucleotide detecting CLTC-TFE3 fusion gene in sample, method and kit
A CLTC-TFE3 and fusion gene technology, which is applied in biological science and biological fields, can solve the problems of poor specificity and high cost, and achieve the effect of simple operation, high accuracy and improved experimental efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] For detecting the oligonucleotide of CLTC-TFE3 fusion gene in the sample, said oligonucleotide comprises: for the upstream and downstream primers and probes of CLTC-TFE3, its base sequence is respectively SEQ ID NO:1,2 and 3.
[0030]Further, the oligonucleotide also includes upstream and downstream primers and probes for the internal reference gene of β-actin, the base sequences of which are SEQ ID NO: 4, 5 and 6, respectively.
[0031] A method for detecting CLTC-TFE3 fusion gene in a sample, said method comprising the following steps:
[0032] (1) extract the RNA in the sample;
[0033] (2) Reverse transcribe the RNA extracted in (1) into cDNA;
[0034] (3) Add the cDNA described in (2) to the reaction tube, and use the upstream and downstream primers and probes for CLTC-TFE3 to detect the fluorescent signal of the CLTC-TFE3 fusion gene in the sample, and its base sequences are SEQ ID NO: 1, 2 and 3; use the upstream and downstream primers and probes for the β-act...
Embodiment 2
[0046] The operating flow of the inventive method:
[0047] (1) Extract tissue RNA from blood: Add 1ml of the erythrocyte lysate in Example 1 to a clean 1.5ml centrifuge tube, take 0.5ml of anticoagulated blood and mix well, let stand at room temperature for 10min; centrifuge at 5000rpm for 5min, discard Supernatant, collect the cells at the bottom; add 0.5ml of the erythrocyte lysate in Example 1 again, centrifuge at 5000rpm for 5min, discard the supernatant, and collect the cells at the bottom; add 1ml TRIzol to the cells, pipette repeatedly until the precipitate is completely dissolved, and stand at room temperature 5min; add 0.2ml chloroform, shake evenly; centrifuge at 14000rpm 4°C for 10min, absorb the supernatant layer and transfer to another new centrifuge tube (do not absorb the white middle layer); add an equal volume of isopropanol, mix well up and down, Let stand at room temperature for 10min; centrifuge at 14000rpm at 4°C for 10min, discard the supernatant, add 1m...
Embodiment 3
[0060] Example 3 Positive plasmid detection and sensitivity detection
[0061] When preparing positive plasmids, first directly synthesize CLTC-TFE3 cDNA, and then insert them into the previously selected plasmid plasmids (here the PMD18-T plasmid is used as an example), so that PMD18-T / CLTC-TFE3 can be prepared positive plasmid.
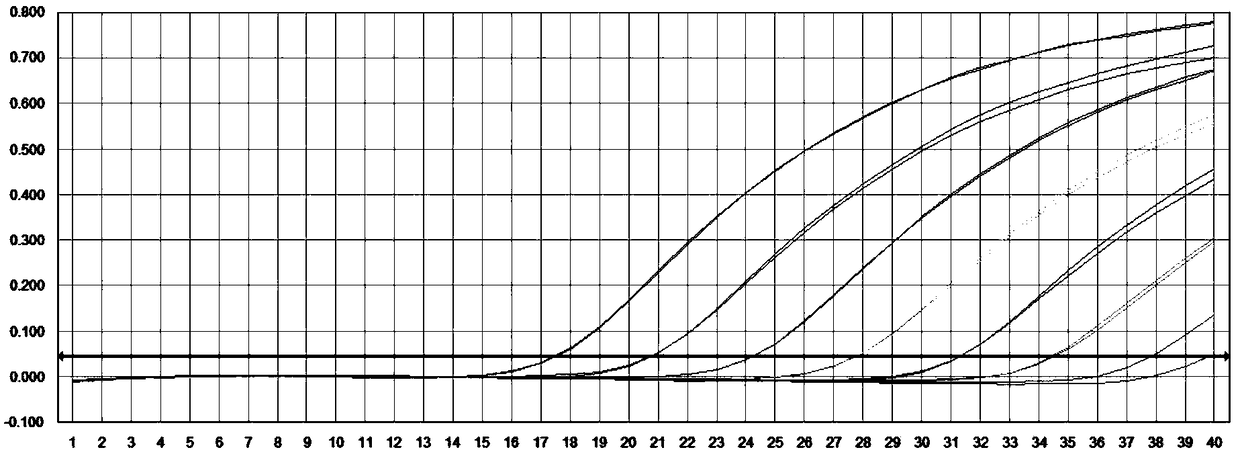
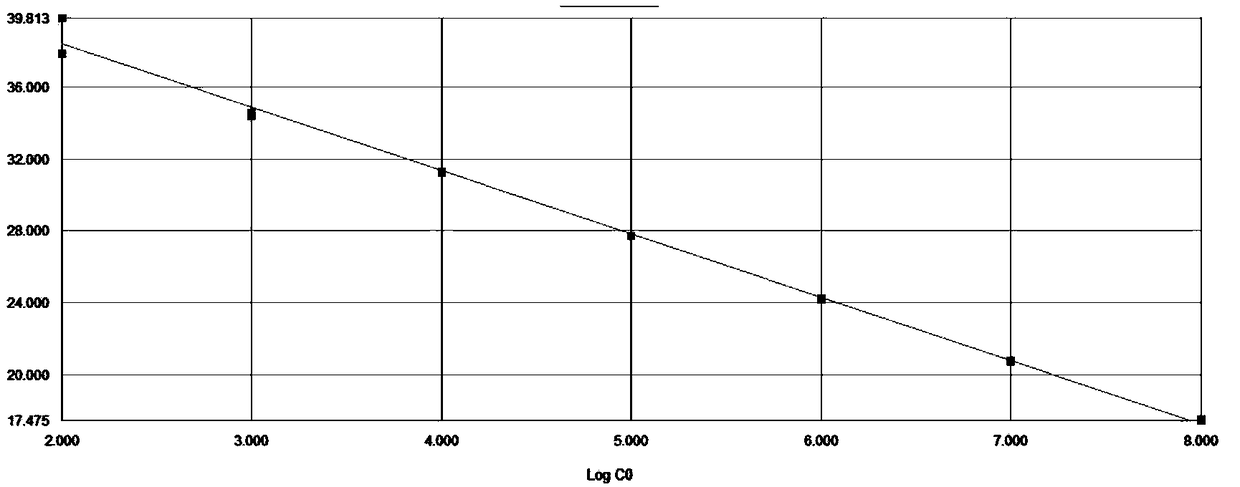
[0062] The prepared PMD18-T / CLTC-TFE3 positive plasmid was serially diluted to obtain a copy number of 10 8 、10 7 、10 6 、10 5 、10 4 、10 3 、10 2 The positive plasmid of copies / μl, and with the positive plasmid of these different concentrations as template, detect according to embodiment 2, the result is as follows figure 1 shown, among them, in figure 1 The positive plasmid concentrations corresponding to each amplification curve in the figure are 10 from left to right 8 、10 7 、10 6 、10 5 、10 4 、10 3 、10 2 copies / uL. Depend on figure 1It can be seen that both CLTC-TFE3 and β-actin are active, and the primers and probes of the present...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com