Quantitative analysis method for epigenetic modification of high-throughput nucleic acid
A technology for epigenetic modification and quantitative analysis, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of high cost of sequencing analysis, large sample damage, etc., to reduce the amount of test samples used and the time used to be short. , the effect of shortening the analysis time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] The establishment of embodiment 1 analytical method
[0085] Take 5'-GAG ACC GGA GTC CGC TTT CCT CTT CCG GAA AAT GTA AGC CGA ACC TAAAGC AAT CAC CAG GG-3' (SEQ ID NO.1) as an example.
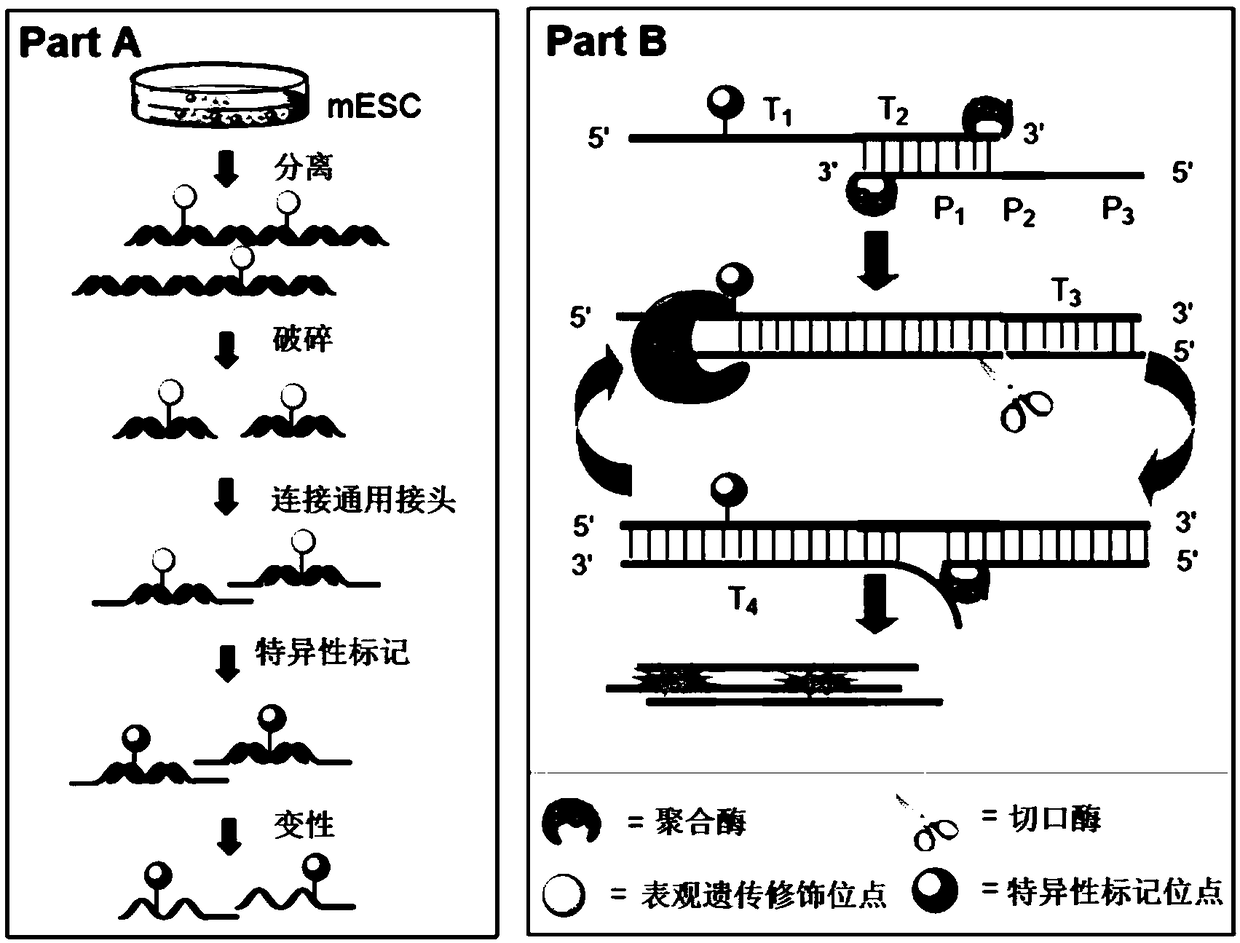
[0086] (1) Preparation of nucleic acid fragments with different epigenetic modifications
[0087] The 8 DNA template sequences are as follows:
[0088] C: 5'-GAG ACC GGA GTC CGC TTT CCT CTT CCG GAA AAT GTA AGC CGA ACC TAAAGC AAT CAC CAG GG-3' (SEQ ID NO.1);
[0089] 5mC: 5'-GAG ACC GGA GTC CGC TTT CCT CTT CCG GAA AAT GTA AGC mCGA ACCTAA AGC AAT CAC CAG GG-3' (SEQ ID NO.2);
[0090] 5hmC: 5'-GAG ACC GGA GTC CGC TTT CCT CTT CCG GAA AAT GTA AGC hmCGA ACCTAA AGC AAT CAC CAG GG-3' (SEQ ID NO.3);
[0091] 5fC: 5'-GAG ACC GGA GTC CGC TTT CCT CTT CCG GAA AAT GTA AGC fCGA ACCTAA AGC AAT CAC CAG GG-3' (SEQ ID NO.4);
[0092] 5caC: 5'-GAG ACC GGA GTC CGC TTT CCT CTT CCG GAA AAT GTA AGC caCGA ACCTAA AGC AAT CAC CAG GG-3' (SEQ ID NO.5);
[0093] A: 5'-GGA CTG GAC TGG ACT GGA CTG GAC TAT CAC CAG G...
Embodiment 2
[0135] Application of embodiment 2 analytical method
[0136] (1) Optimization of reaction conditions for isothermal amplification of total DNA from actual samples
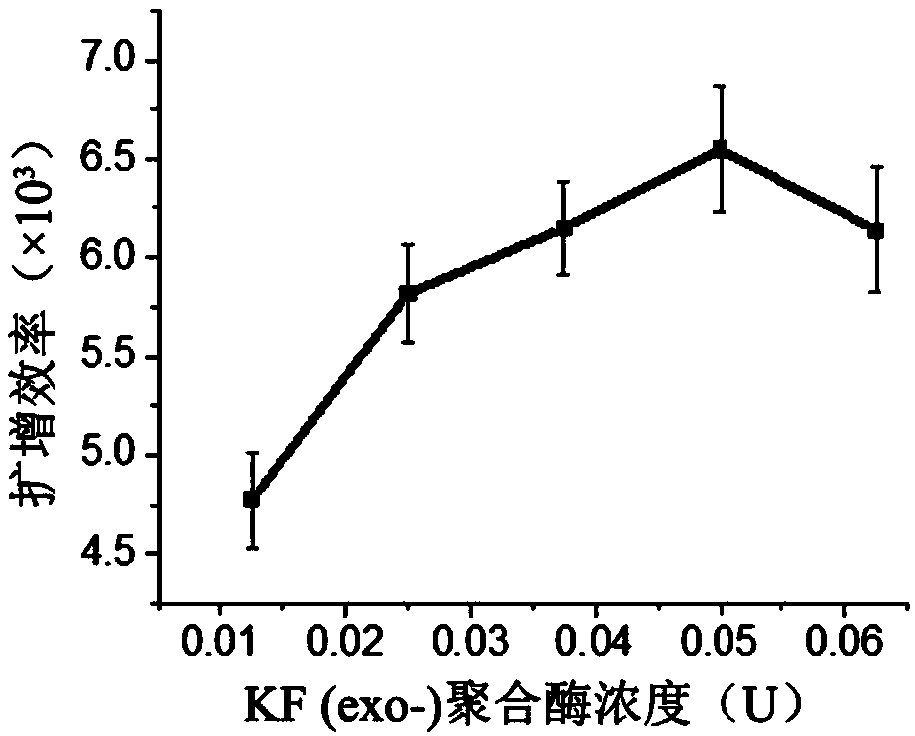
[0137] Optimization of conditions for total DNA methylation assay in real samples by single factor rotation method
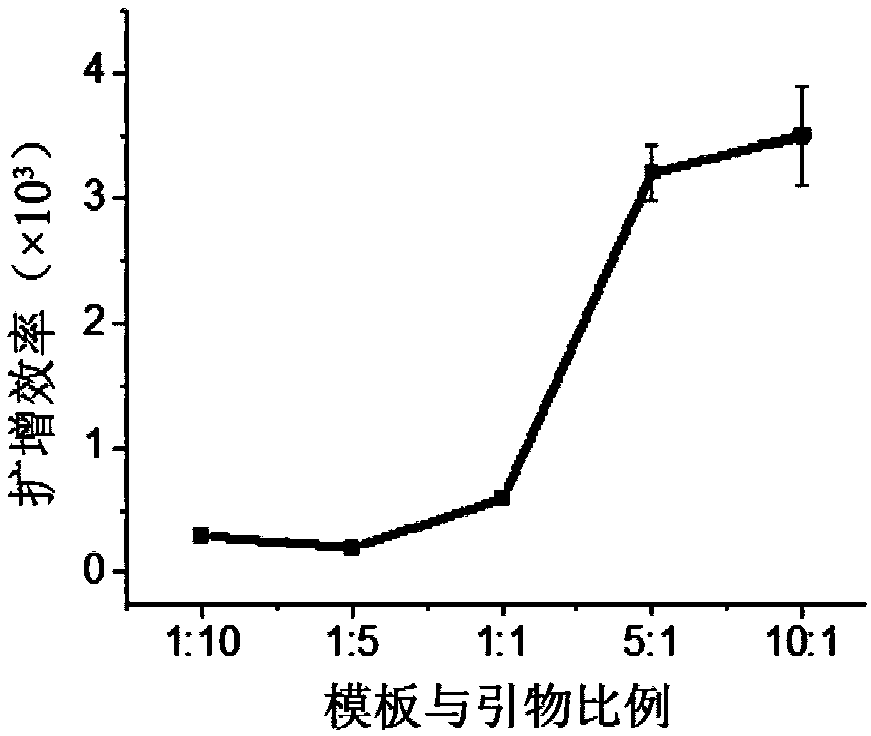
[0138] (a) template usage
[0139] Fix the amount of primer (100nM), prepare DNA fragment solutions with different concentrations, and optimize the optimal template amount. The reaction was prepared in two parts, Part A and Part B. Part A included nicking enzyme buffer (1×), dNTPs (250 μM), primer strands and template solutions of different concentrations, and Part B included polymerase buffer (1× ), SYBR Green II fluorescent dye (2×), KF (exo-) polymerase, Nt.BsmAI Nease nickase, all the above operations were performed on ice. Part A and Part B were mixed, and quickly placed in a quantitative PCR instrument for amplification reaction at 37°C. The reaction concentrations of DNA were 81.75, 65.40...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com