Multi-gene polymerization effect analytic breeding method for increasing lambing number of goats
A litter size and multi-gene technology, which is applied in biochemical equipment and methods, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems of long time-consuming, poor selection effect of single gene molecular markers, low breeding efficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
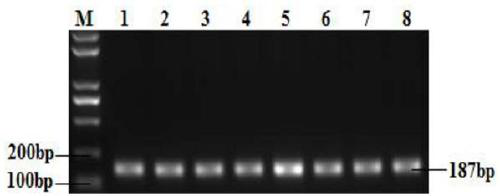
[0076] The establishment of the PCR-RFLP detection method of embodiment 1 GnRHR gene
[0077] (1) Primer design
[0078] According to the goat GnRHR gene sequence (Gene ID on NCBI: 100860755), utilize Primer5.0 software to design a pair of primers, sequence is as follows:
[0079] GnRHR:
[0080] SEQ ID NO.4: Upstream primer PCR-F: 5'-TCTTGAAGCTGTATCAGCCATA-3';
[0081] SEQ ID NO.5: Downstream primer PCR-R: 5'-GTGTTGAAAATTGTGGAGAGTAGA-3'.
[0082] (2) PCR amplification system and program setting
[0083] PCR reaction system: 10 μL system contains 1 μL of genomic DNA template, 0.2 μL of upstream and downstream primers, 5 μL of TaqDNA polymerase, supplemented with deionized double-distilled water to the final volume.
[0084] PCR reaction program: pre-denaturation at 94°C for 2 min, 35 cycles (denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 30 s), extension at 72°C for 5 min, and storage at 4°C.
[0085] The PCR reaction product was detected...
Embodiment 2
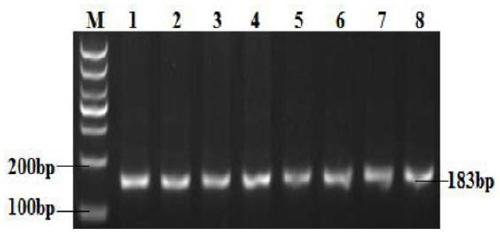
[0102] The establishment of the PCR-HRM detection method of embodiment 2 FSHR gene
[0103] (1) Primer design for high resolution melting curve (HRM)
[0104] According to the goat FSHR gene sequence (Gene ID on NCBI: 100861291), utilize Primer5.0 software to design a pair of primers, sequence is as follows:
[0105] FSHR:
[0106] SEQ ID NO.6: Upstream primer PCR-F: 5'-TACCAGCTCCCAACGCAGAC-3';
[0107] SEQ ID NO.7: Downstream primer PCR-R: 5'-GACAGAGTCGATGGTGGCAT-3'.
[0108] (2) High resolution melting curve (HRM) reaction conditions
[0109] Use 55-65°C temperature gradient PCR to understand the optimal annealing temperature of the primers to obtain specific PCR products and improve the sensitivity of high-resolution melting.
[0110] PCR reaction system (10 μL): DNA template 1 μL, upstream and downstream primers 0.3 μL each, Dye 5 μL, RNase-free water 3.4 μL.
[0111] The PCR reaction program was: pre-denaturation at 94°C for 2 min, 40 cycles (denaturation at 94°C for...
Embodiment 3
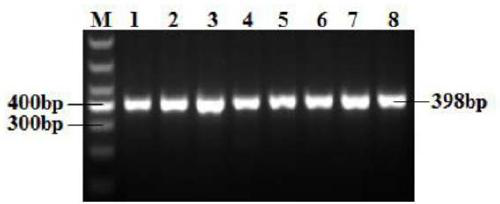
[0129] The establishment of the PCR-RFLP detection method of embodiment 3 BMP6 gene
[0130] (1) Primer design
[0131] According to the goat BMP6 gene sequence (Gene ID on NCBI: 100860793), utilize Primer5.0 software to design a pair of primers, sequence is as follows:
[0132] BMP6:
[0133] SEQ ID NO.8: Upstream primer PCR-F: 5'-TGTGCCCTCACCTGCTGTCTC-3';
[0134] SEQ ID NO.9: Downstream primer PCR-R: 5'-CCCGGCCTTCCTCCTTTAACTC-3'.
[0135] (2) PCR amplification system and program setting
[0136] PCR reaction system: 10 μL system contains 1 μL of genomic DNA template, 0.2 μL of upstream and downstream primers, 5 μL of TaqDNA polymerase, supplemented with deionized double-distilled water to the final volume.
[0137] PCR reaction program: pre-denaturation at 94°C for 2 min, 35 cycles (denaturation at 94°C for 30 s, annealing at 59°C for 30 s, extension at 72°C for 30 s), extension at 72°C for 5 min, and storage at 4°C.
[0138] PCR reaction product is detected with 2% ag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com