Enzymatic preparation of beta alanine
An alanine and amino acid technology, applied in the directions of enzymes, lyases, carbon-carbon lyases, etc., can solve the problems of environmental pollution of nitrile substances and complicated product purification steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1 Aspartic Acid Decarboxylase Genetic Engineering Bacteria Construction
[0042] Synthesize the sequence SEQ ID NO:3 through the whole gene sequence, design restriction sites NdeI and BamHI at both ends, clone into the corresponding sites on the pSH plasmid, obtain the recombinant plasmid pSH-Pandcg, and then transform it into E. coli for expression by calcium chloride method In the host BL21 (DE3) competent cells, coat the LB medium plate containing kanamycin, cultivate overnight at 37°C, pick a single colony, inoculate it into a test tube containing LB medium, cultivate overnight, collect the bacteria by centrifugation, and extract The plasmid was extracted, the gene sequence was confirmed to be correct, and the recombinant genetically engineered strain expressing the wild-type Pandcg enzyme was obtained.
Embodiment 2
[0043] Example 2 Error-prone PCR method constructs aspartate decarboxylase (Pandcg) random mutation point library
[0044] Using SEQ ID NO:3 as a template, an error-prone PCR technique was used to construct a random mutant library.
[0045] Forward primer Pandcg-Nde-F: 5'-ATGTACCTGCGCACCATCCTCGGAAG-3',
[0046] Reverse primer Pandcg-Xho-R: 5'-CTCGAGCTAAATGCTTCTCGACGTCAAAAGC-3'.
[0047] 50μL error-prone PCR reaction system includes: 50ng plasmid template pSH-Pandcg, 30pmol pair of primers Pandcg-Nde-F and Pandcg-Xho-R, 1×Taq buffer, 0.2mM dGTP, 0.2mM dATP, 1mM dCTP, 1mM dTTP, 7mMMgCl 2 , (0mM, 0.05mM, 0.1mM, 0.15mM, 0.2mM) MnCl 2 , 2.5 units of Taq enzyme (fermentas). The PCR reaction conditions were: 95°C for 5 minutes; 94°C for 30s, 55°C for 30s, 72°C for 2min / kbp; 30 cycles; 72°C for 10min. The 2.0kb random mutation fragment was recovered from the gel as a large primer, and MegaPrimer PCR was performed with KOD-plus DNA polymerase: 94°C for 5min; 98°C for 10s, 60°C for...
Embodiment 3
[0048] The high-throughput screening of embodiment 3Pandcg mutant library
[0049] 3.1 Select the transformant in the mutant library, inoculate it into a 96-well deep-well culture plate containing 700 μg / mL of LB medium, the medium contains 100 μg / mL kanamycin, culture at 37°C for 6 hours, and add a final concentration of 0.1 mM IPTG , cooled to 25°C, and incubated overnight. Centrifuge at 5000rpm for 10min, discard the supernatant, freeze at -70°C for 1h, and thaw at room temperature for 30min. Add 200 μL of 0.1M potassium phosphate buffer (pH 8.0) to resuspend the bacteria for the determination of Pandcg enzyme activity.
[0050] 3.2 Determination and screening of mutants with high enzyme activity
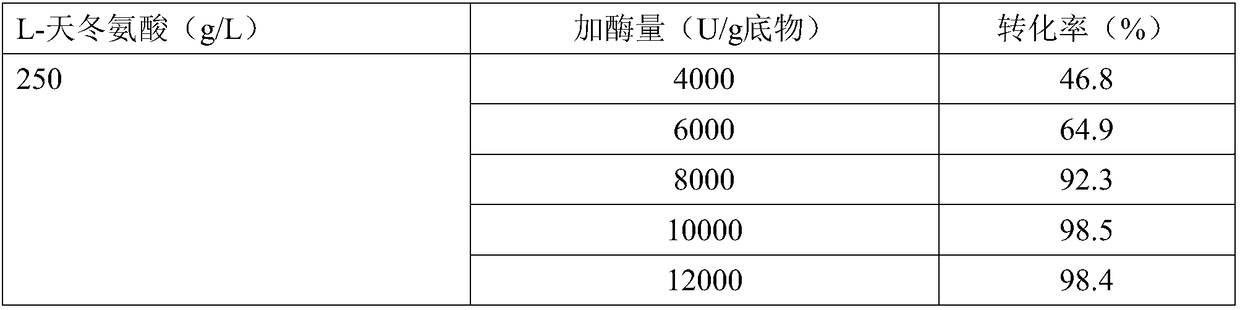
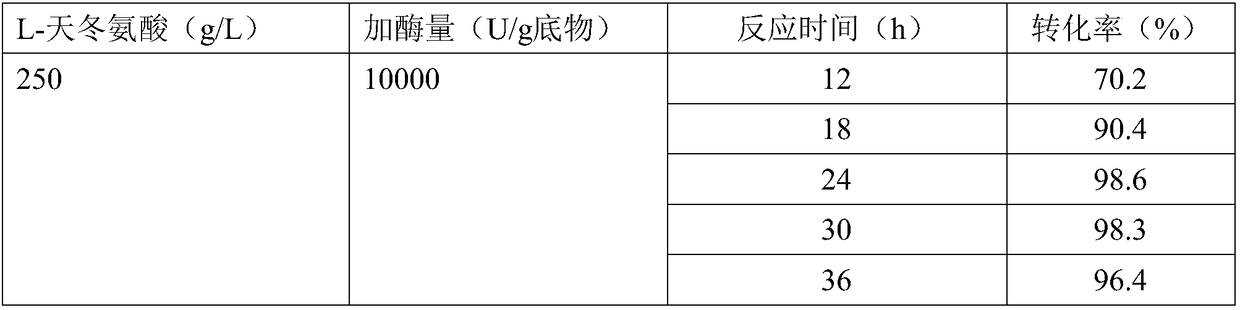
[0051] Substrate reaction solution: 200 g / L L-aspartic acid, adjusted pH with NaOH to dissolve.
[0052] Termination reaction solution: 0.5ml of 1M NaOH solution.
[0053] Homogenize 80 μL of the cell suspension in step 3.1 above to break the wall, add 80 μL of the substrate ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com