Transposase high-activity mutant in halophilic archaebacteria
A high-activity, mutant technology, applied in the field of molecular biology, can solve the problems of high price of transposase, increase of transposase purification cost, etc., achieve good tolerance and reduce purification cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0014] Example 1 Construction of transposase highly active mutant Escherichia coli engineering bacteria
[0015] The highly active mutation W64K was introduced into the transposase (GenBank: KXA97509.1) gene of the non-culturable microbial halophilic archaea SCGC-AAA259I14, and the codon-optimized gene sequence was codon-optimized with reference to the codon preference of Escherichia coli. The sequence was synthesized by Jinweizhi Company and constructed into the pTYB4 expression vector to obtain the pTYB4W64K plasmid
[0016] Transform the pTYB4W64K plasmid into Escherichia coli ER2566 expression strain, use LB medium, cultivate to OD at 37 degrees 600 =0.6, add 0.1mM IPTG, lower the culture temperature to 23°C, continue to culture for 6h, and collect the cells by centrifugation.
Embodiment 2
[0017] Example 2 Purification of Transposase High Activity Mutants
[0018] Take 25-30 g of Escherichia coli cells expressing transposase highly active mutants, and add 500 mL of buffer A (20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 10% Glycerol) at a ratio of 1:17-20 , after mixing, perform homogeneous bacteriostasis (1000bar / 2 times), after bacteriostasis, centrifuge the sample (14000 rpm, 20 min, 4°C), keep the supernatant and discard the precipitate after centrifugation.
[0019] Slowly add the centrifuged supernatant to a 5 mL Chitin column; after all the destructed supernatant passes through the Chitin column, use buffer A to wash the column for 5-7 times the column volume to ensure that some non-specific binding proteins in the column are washed out. At this time, the highly active transposase mutant is bound to the Chitin filler.
[0020] Weigh the DTT solid and dissolve it in 20 mL buffer A (the final concentration of DTT is 50 mM), slowly add the above buffer solution to the...
Embodiment 3
[0022] Example 3 Application of Transposase High Activity Mutants in the Field of Molecular Biology
[0023] Application of transposase highly active mutants in next-generation sequencing library construction:
[0024] 3.1 Construction of transposomes (Transposome)
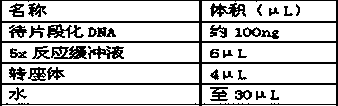
[0025] 3.1.1 Prepare the following reaction system:
[0026]
[0027] 3.1.2. Mix the reactants evenly and react at 25°C for 30 minutes.
[0028] 3.1.3. After the reaction, the transposomes can be used for subsequent reactions immediately, or stored at -20°C.
[0029] 3.2 Fragmentation reaction (Tagmentation)
[0030] 3.2.1. Prepare the following reaction system
[0031]
[0032] 3.2.2. React at 37°C for 2 hours or at 56°C for 10-15 minutes.
[0033] 3.3 After purification, nick translation, PCR enrichment, purification and other steps, the product obtained in the previous step can be sequenced on the machine.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com