Application of SNP (Single Nucleotide Polymorphism) sites of yak whole genome, primer set for detecting and kit
A whole genome and primer set technology, applied in the field of animal husbandry, achieves the effects of easy standardization and automatic detection, and high determination accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] This example provides a method for selecting the site with the highest reliability of the SNP mutation from the resequencing data of 96 yaks to prepare a chip.
[0038] First, the Illumina X Ten sequencing platform was used to perform whole-genome resequencing data (10X coverage for each individual) from 32 different local yak populations (3 individuals in each population, a total of 96 yak individuals) from all over China, and 2592G the sequencing data. On this basis, a quality control is first performed to filter out data with low sequencing quality. Next, we use the BWA comparison software to compare the total reads to the reference genome, generate a comparison result file in sam format, and then use samtools to convert the result file in sam format into bam format and sort it. Finally, we use the identification SNP most prevalent in GATK to identify SNP mutations in 96 yaks. Finally, we selected some high-confidence sites from the 96 SNP mutations of yaks to make...
Embodiment 2
[0051] This example provides a screening method for 268 SNP sites of yak meat production-related traits.
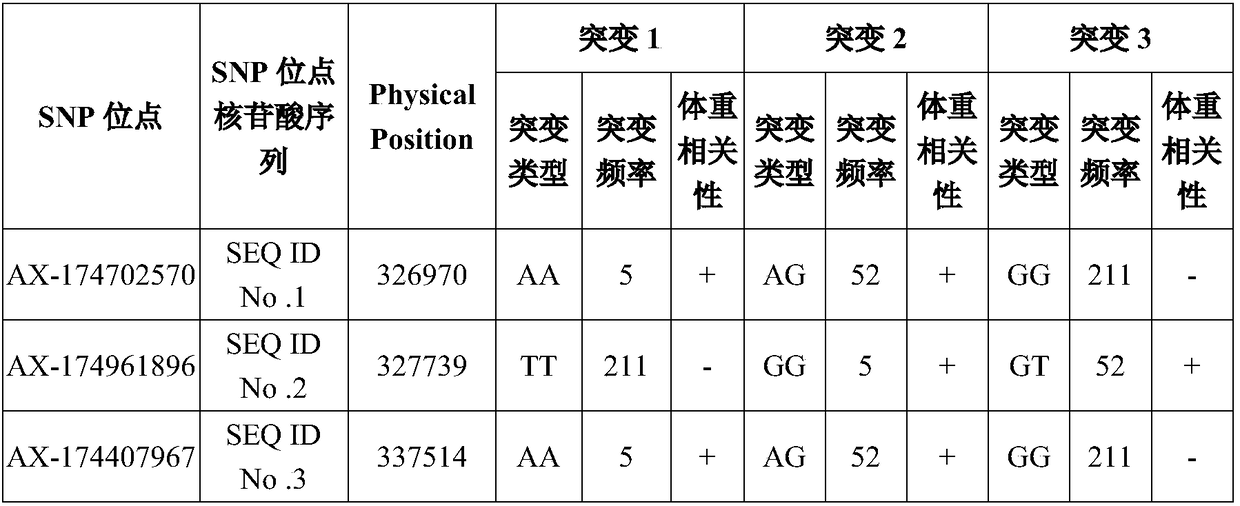
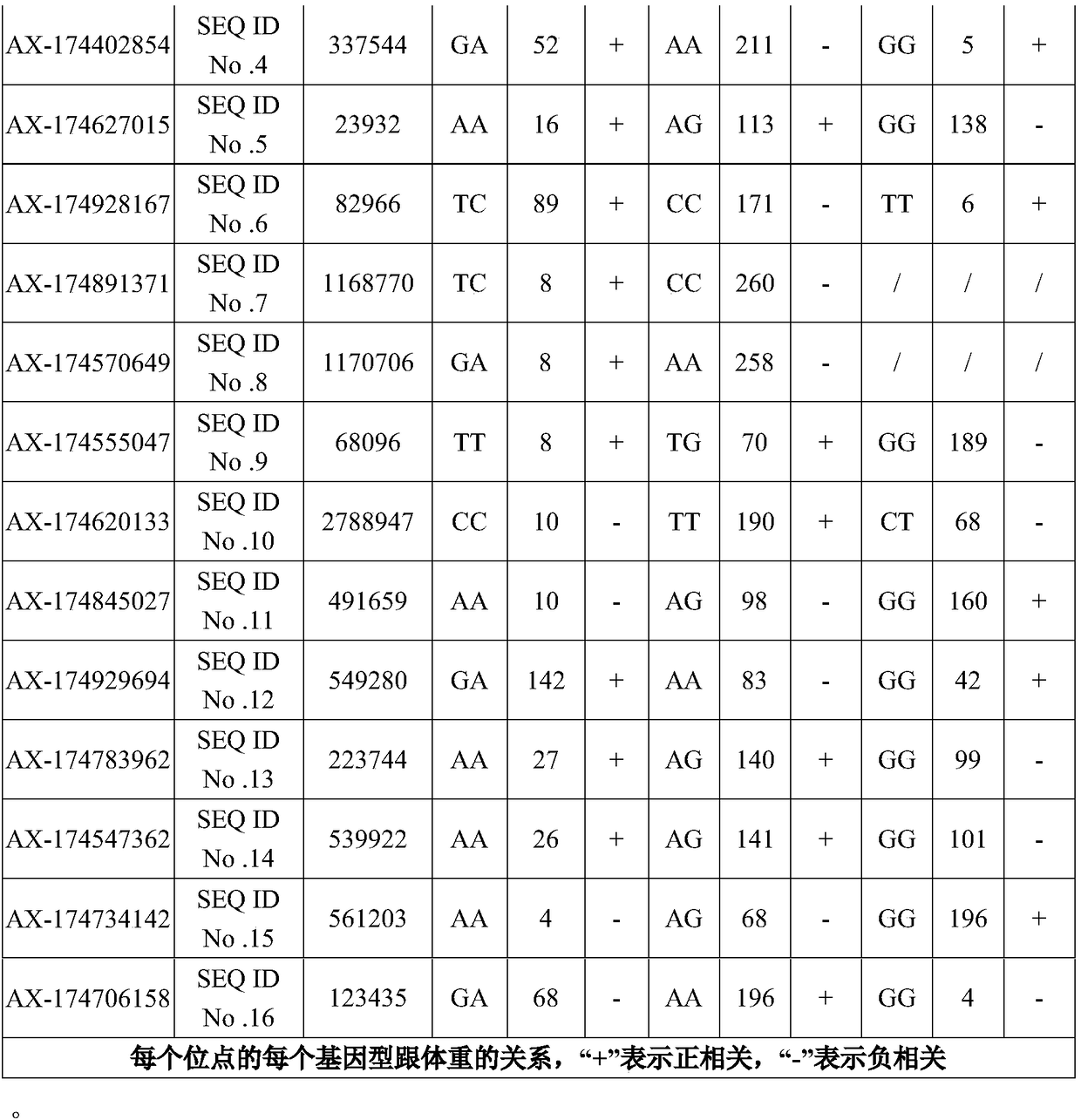
[0052] A total of 268 individuals from 3 nationally certified yak populations, including Jiali yak, Pali yak and Sibu yak, were selected, and SNP sites were screened by the chip prepared in Example 1. The samples were strictly selected yak individuals aged 4-9 in the young adults in each group, and healthy individuals with low body weight and high body weight were screened in each group as samples for microarray screening. The body weight data of each individual was recorded in detail, and GWAS analysis was performed based on the obtained chip screening data, and finally 16 loci most related to yak meat production were found.
[0053] The specific steps are:
[0054]Sample gDNA was quantified using NanoDrop ND-2000 (Thermo Scientific) and DNA integrity was detected by gel electrophoresis. After passing the DNA quality inspection, sample amplification, fragmentation, pre...
Embodiment 3
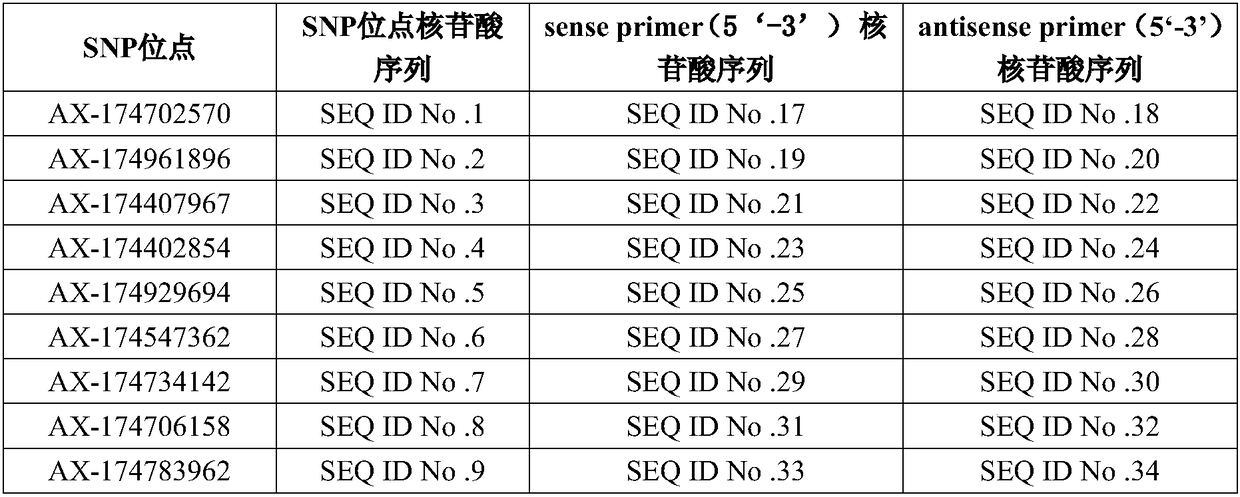
[0061] This embodiment provides a primer set for detecting the 16 SNP sites screened in Example 1, and the corresponding relationship between the nucleotide sequence of the primer set and the SNP site is shown in the following table:
[0062]
[0063]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com