Construction method of IPEC-J2 cell with APN gene knockout
A technology of IPEC-J2 and construction method, which is applied in the field of construction of porcine intestinal epithelial cell line (IPEC_J2) model, and can solve problems such as not yet obtained
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
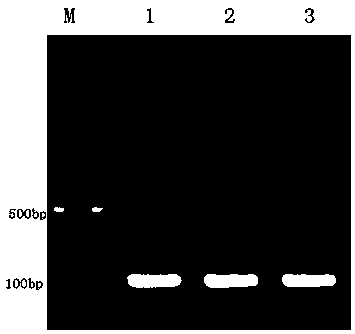
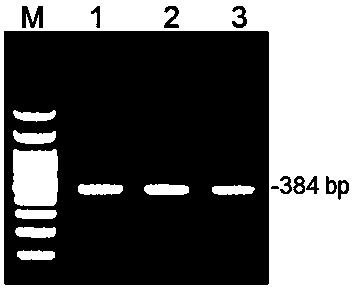
[0028] Target site design and sgRNA sequence synthesis:
[0029] The CDs sequences of all transcripts of the porcine APN gene (Gene ID: 397520) were obtained according to the NCBI database, and the first exon of the CDs region was found for target site design. Use the online tool CRISPR Design (http: / / crispr.mit.edu / ) to design the sgRNA guide sequence:
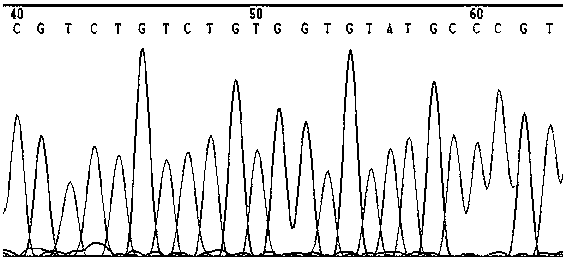
[0030] 5'-TCTGTCTGTGGTGTATGCCC-3', in which the sgRNA action site is located in the first exon of the porcine APN gene, and CACCG is added to its 5' end to form a positive strand Oligo DNA.
[0031] At the same time, the reverse complementary sequence of the sgRNA sequence was synthesized: 5'-GGGCATACCACCAGACAGA-3', and AAAC was added at the 5' end to form a negative strand Oligo DNA.
[0032] The result is as follows:
[0033] Positive strand Oligo DNA: 5'-CACCGTCTGTCTGTGGTGTATGCCC-3';
[0034] Negative strand Oligo DNA: 5'-AAACGGGCATACCACCAGACAGA-3'.
[0035] . Construction of targeting vector:
[0036] (1) Anneal the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com