Method for quantitative evaluating HP DNA hairpin configuration on substrate surface based on enzymic hydrolysis ability and background signal eliminating method based on enzymic hydrolysis ability
An evaluation method and background signal technology, applied in the field of DNA self-assembled membranes, can solve the problem of inability to distinguish the number and ratio of different configurations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0052] 1. Removal of dimers in the Hairpin-16 / ssDNA self-assembled membrane
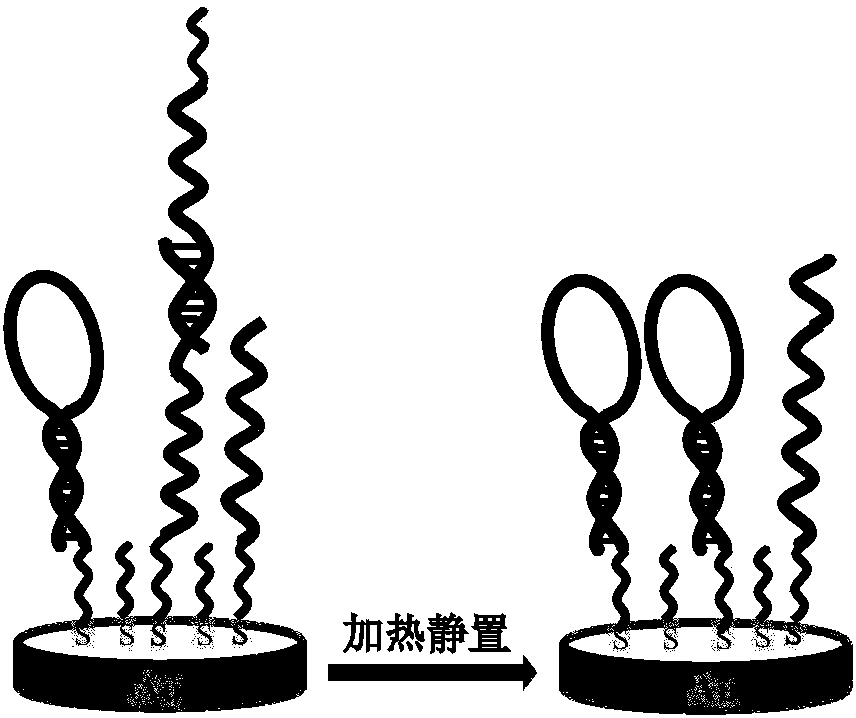
[0053] (1) Construction of Hairpin-16 / ssDNA self-assembled membranes (SAMs): 0-1 μmol Hairpin-16 and ssDNA were mixed and mixed with a vortex shaker to prepare Hairpin-16 / ssDNA mixed solutions with different ratios. Soak the activated gold electrode in the pre-configured Hairpin-16 / ssDNA mixed solution, and place it at room temperature for 4-24h to obtain the Hairpin-16 / ssDNA self-assembled membrane, as shown in figure 1 shown.
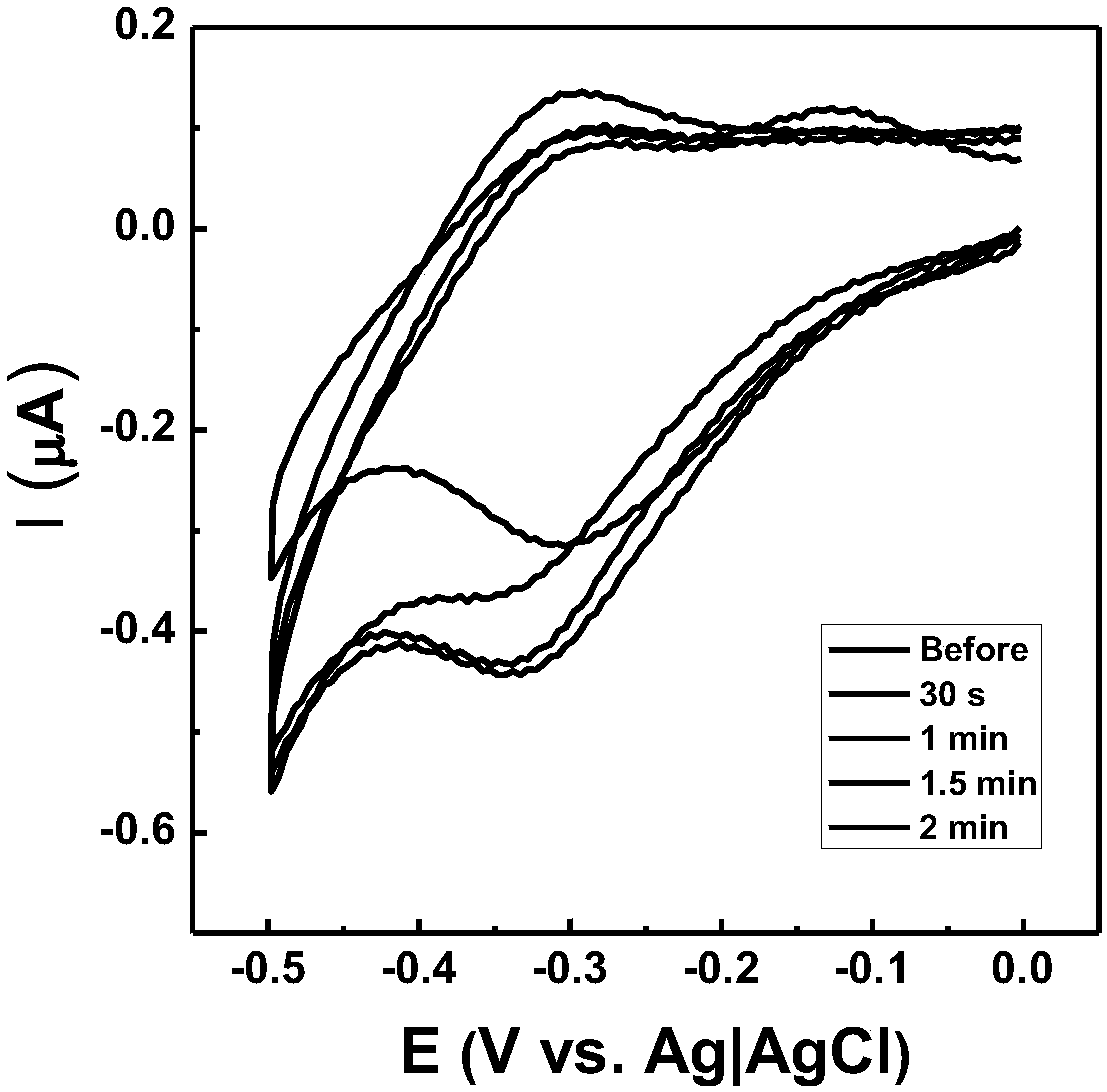
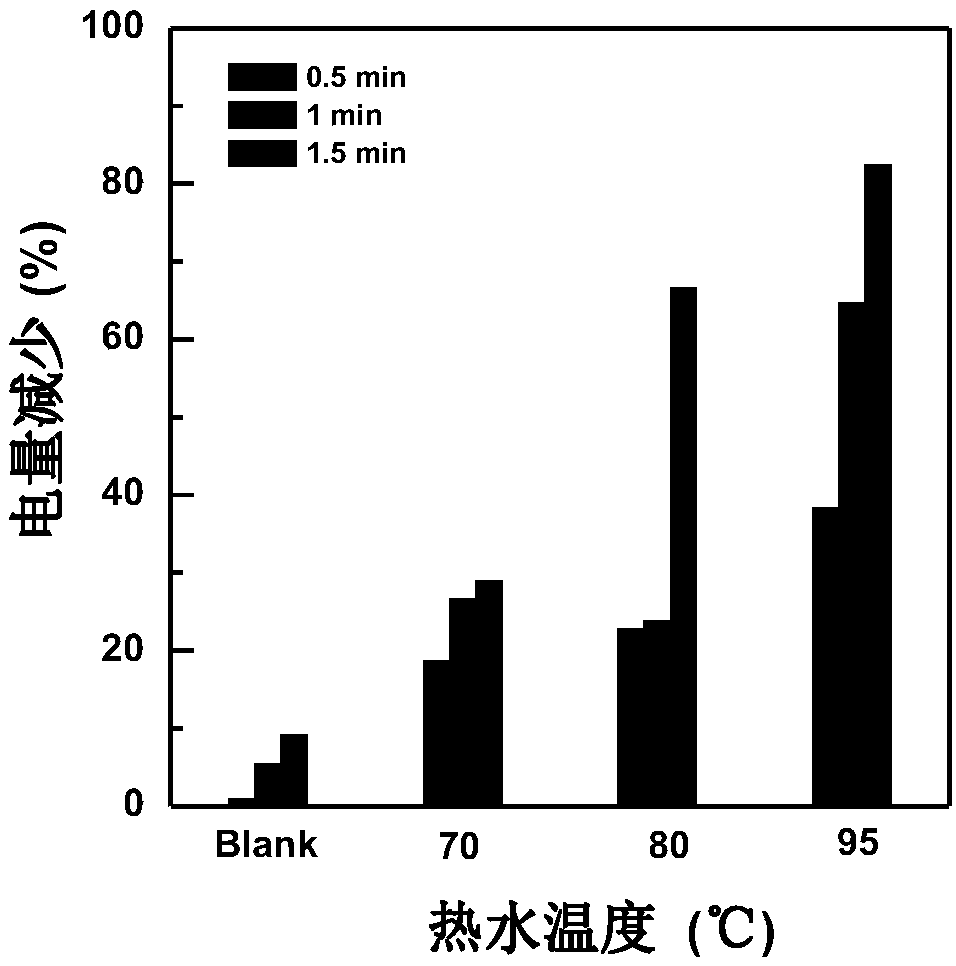
[0054] (2) Removal of dimers in the Hairpin-16 / ssDNA self-assembled membrane: Place the above-mentioned gold electrode in newly prepared hot water at 75-95°C for denaturation for 0.5-3 minutes, and then place the electrode statically in 10mM Tris-buffer (100mM~200mM Na + , 30mM~80mM Mg 2+ ) for 1 to 12 hours, the dimer configuration on the electrode surface can be removed, and the hairpin configuration can be refolded. Such as figure 2 As shown in the cyclic voltammogr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com