Molecular marker of upland cottonseed oil content gene GhKAS1 and screening method thereof
A technology of molecular markers and screening methods, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problem of lack of genetic information of cottonseed oil, etc., to achieve clear location, convenient identification, convenient and fast detection Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] The discovery of GrKAS1 gene in embodiment 1 Raymond's cotton
[0022] Using the genes related to fatty acid synthesis in the Raymond cotton genome as a reference sequence, 162 pairs of Indel primers were designed for genotype analysis, and the cotton kernel oil content of 503 upland cotton varieties in two years and four environments was used in TASSEL2.1 Based on the linear model, a gene related to cottonseed oil content was detected as the predicted KAS1 gene in Raymond cotton, named GrKAS1. Specifically, 162 pairs of Indel primers were used to carry out PCR amplification using 503 upland cotton varieties as templates. The results of electrophoresis and polyacrylic gel electrophoresis were sorted out and correlated, and it was found that the gene amplified by PCR with a pair of Indel primers was related to oil content, and the marker was named GrKAS1. The pair of Indel primers corresponds to the predicted GrKAS1 gene in Raymond cotton, and the results of the associat...
Embodiment 2
[0026] Example 2 Molecular marker of cotton kernel oil content gene GhKAS1 in upland cotton
[0027] 1. Use CottongenBlast to compare the gene to the genome database of upland cotton, find out the fragments with high homology, and determine its position on the chromosome. It can be determined that GrKAS1 has 6 copies in the upland cotton genome, which is 6 candidates Gene.
[0028] (1) Use the website http: / / www.cottongen.org / to find out the copy number and physical location of GrKAS1 in the upland cotton genome.
[0029] (2) Predict the gene structure and protein structure of 6 candidate genes, the website used is http: / / meme.ebi.edu.au / meme / cgibin / meme.cgi.
[0030] 2. Design specific primers for 6 candidate genes
[0031] (1) Find the predicted sequence of the GhKAS1 gene in the A genome, D genome, sea island cotton and upland cotton TM-1 (compare and predict whether it is the KAS1 gene in NCBI), and design primers to amplify the full length.
[0032] (2) Compare the am...
Embodiment 3
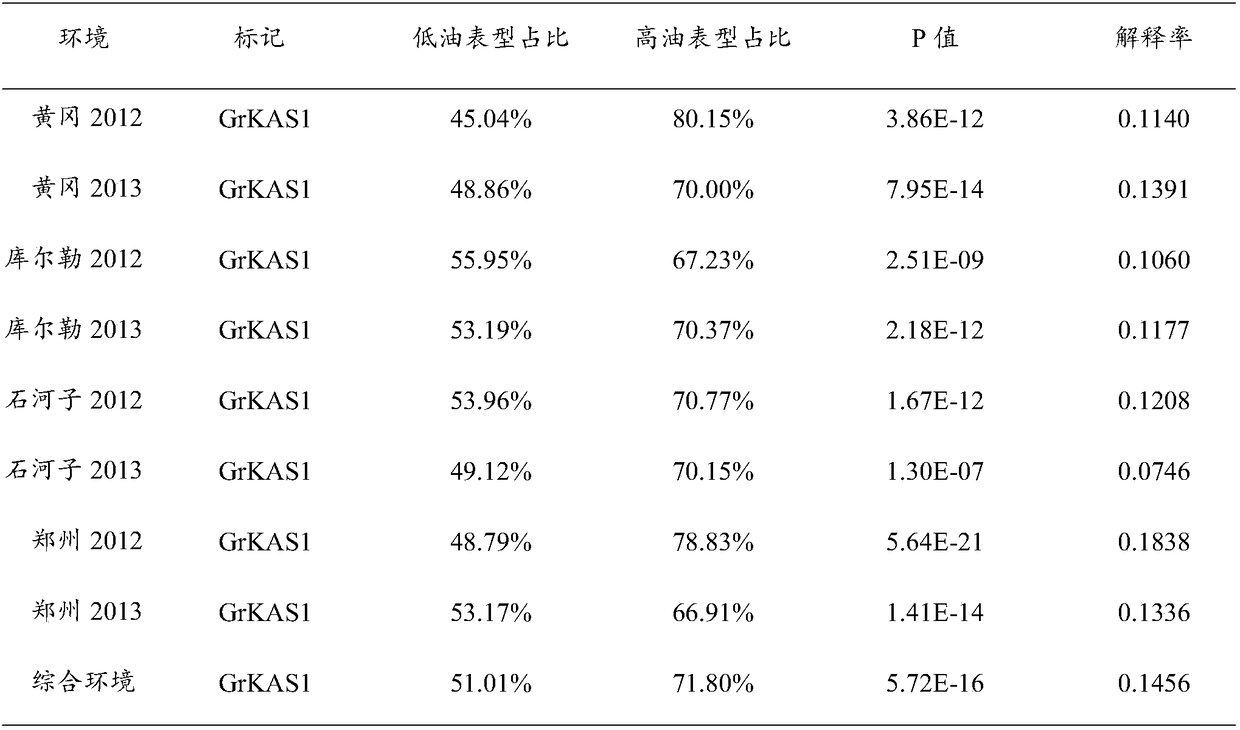
[0041] Example 3 Association analysis of molecular marker mutation sites of cotton kernel oil content gene GhKAS1 in upland cotton
[0042] 1. Extreme material resequencing to determine the mutation site
[0043] From 503 upland cotton varieties, 10 cotton kernels with extreme oil content were selected to extract DNA, among which 10 cotton kernels with extreme oil content included 5 low-oil materials and 5 high-oil materials, and then 10 cotton kernels with extreme oil content were selected. The GhKAS1 gene in each extreme material was resequenced, and the resequencing data were processed with DNAMANV6 software. Compare sequence differences, identify mutation sites, and determine mutation sites;
[0044] Specifically: extract the DNA of 10 extreme material seeds, and use this as a template to amplify the GhKAS1 gene. Because the GhKAS1 gene is too long, primers were designed to amplify in segments, and then TA cloned. The system for TA cloning is 5 microliter system: 2.5 mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com