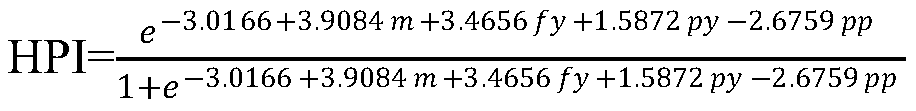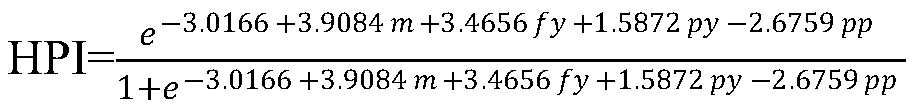A Breeding Method for Simplified Selection of High-performance A2A2 Homozygous Genotype Dairy Cows Based on Pedigree Information
A production performance and information simplification technology, applied in informatics, genomics, evolutionary biology, etc., can solve problems such as heavy workload and inability to take into account the high-yield performance of dairy cows, and achieve the effect of reducing workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Analysis of the breeding value of different genotypes of β-casein and production traits of bulls
[0049] 1. Data collection
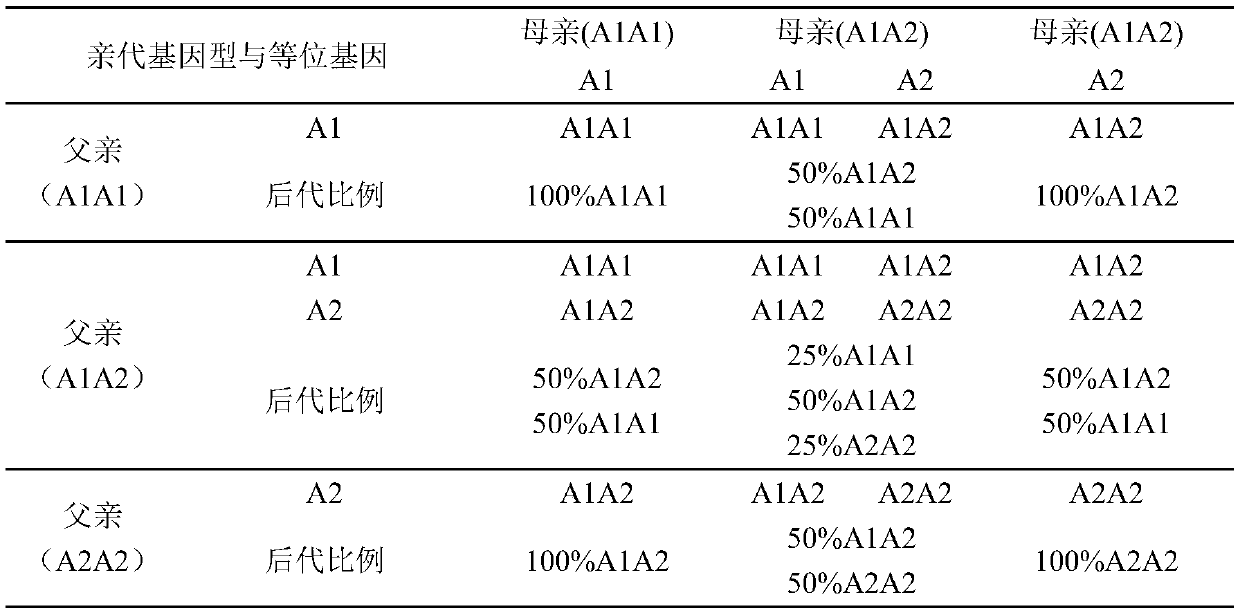
[0050] The bull genetic evaluation and pedigree information data of more than 600,000 cattle were downloaded from the Canadian CDN, among which 3881 bulls currently in use for breeding have all undergone β-casein genotype identification. The cattle number, pedigree, genotype and breeding values of these bulls such as milk yield, milk fat amount, milk protein amount, milk fat percentage, milk protein percentage and other traits directly related to production performance were recorded and established to establish a database.
[0051] 2. Statistics
[0052] Using the SAS software GLM process to analyze the β-casein allele and genotype frequency in the bull population and its impact on production performance, it was found that the A1 and A2 allele frequencies were 39.69% and 60.31%, respectively, and the A1A1, A1A2 and A2A2 genes The g...
Embodiment 2
[0077] Embodiment 2 HPI index method compares with PCR molecular detection method
[0078] The inventor of the present invention selected a total of 151 individual fathers whose fathers were A1A2 / A2A2 among about 500 adult cows participating in DHI large-scale cows around Jinan, Shandong, collected relevant data, collected milk samples and measured milk components, and utilized the method of the present invention Calculate the HPI index of the cattle, and screen the cattle according to the principle of HPI>0.85; at the same time, collect blood samples, extract DNA, and refer to the patent CN201610260677.8 method for genotype determination to obtain individual genotypes.
[0079] Specific steps:
[0080] (1) Collect milk samples from the test cow group (151 heads), and record the calving date, sampling date, and milk production of individual cows. For milk sample sampling method, refer to "Technical Specifications for Production Performance Measurement of Chinese Holstein Catt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com