A kind of cell line for detecting hepatitis A virus titer and its construction method and application
A technology of hepatitis A virus and its construction method, which is applied in the field of cell lines and its construction for qualitative and quantitative detection of hepatitis A virus titer, which can solve complex and cumbersome operations, long time-consuming detection of hepatitis A virus infectivity titer, etc. Problems, to achieve good repeatability, shorten the time of marketing immunization, and ensure the quality of the effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Step (1), plasmid construction:
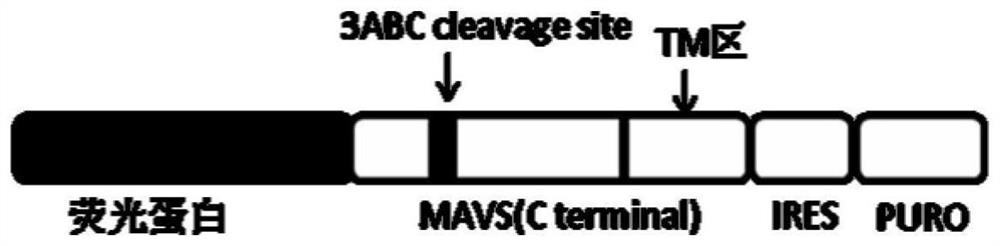
[0077] Using the total RNA in Huh7.0 cells as a template, using P1 and P2 as primers, RT-PCR was used to amplify the MAVS C-terminal gene sequence MAVS (C396-540), and after double digestion with BsrGI / MluI, it was inserted into the The LV-EGFP-MAVS(C396–540)-IRES-PURO-WPRE recombinant plasmid was constructed in the enzyme-treated lentiviral expression vector LV-EGFP-IRES-PURO-WPRE vector. After sequencing analysis, the sequence was correct and it was stored in Standby at -20°C.
[0078] The specific method is as follows:
[0079] PCR amplification system and amplification program
[0080] 1. Synthesize cDNA
[0081] Use takara's PrimeScriptII 1st Strand cDNA synthesis Kit to synthesize cDNA according to the instructions:
[0082] system:
[0083] Volume (ul) Huh7.0 RNA 2 Primer P2 0.5 dNTP 4 RNase free ddH 2 o
6.5ul Total 13
[0084] 65°C for 5 minutes, followed by a rapid ice bat...
Embodiment 2
[0116] Step (1), plasmid construction:
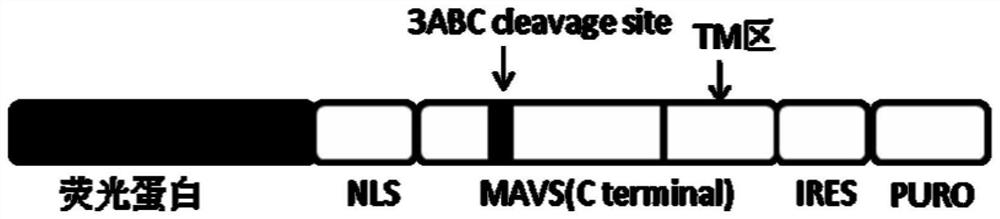
[0117] Using the total RNA in Huh7.0 cells as a template, using P2 and P3 as primers, RT-PCR was used to amplify the MAVS C-terminal gene sequence hMAVS (C396-540), and after double digestion with BsrGI / MluI, it was inserted into the The LV-mCherry-NLS-hMAVS(C396-540)-IRES-PURO-WPRE recombinant plasmid was constructed in the enzyme-treated lentiviral expression vector LV-mCherry-IRES-PURO-WPRE vector. After sequencing analysis, the sequence was correct and the Store at -20°C for later use. The structure of the constructed mcherry-NLS-hMAVS(C396-540)-IRES-PURO-WPRE is shown in Figure 7 .
[0118] 1. Synthesize cDNA
[0119] Use takara's PrimeScriptII 1st Strand cDNA synthesis Kit to synthesize cDNA according to the instructions:
[0120] system:
[0121] Volume (ul) Huh7.0 RNA 2 Primer P2 0.5 dNTP 4 RNase free ddH 2 o
6.5ul Total 13
[0122] 65°C for 5 minutes, followed by a rapid ...
Embodiment 3
[0153] Step (1), plasmid construction:
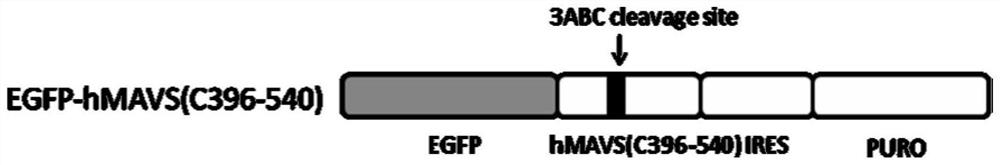
[0154] The total RNA of the tree shrew liver was used as a template, and P4 and P5 were used as primers to amplify the MAVS C-terminal gene sequence MAVS (C365-503) by RT-PCR. After double digestion with BsrGI / MluI, insert it into the LV-EGFP-tMAVS(C365–503)-IRES-PURO-WPRE recombinant plasmid was constructed in the lentiviral expression vector LV-EGFP-IRES-PURO-WPRE vector. After sequencing analysis, the sequence was correct and it was stored at -20 ℃ for later use. The structure of the constructed EGFP-tMAVS(C365-503)-IRES-PURO-WPRE is shown in Figure 11 .
[0155] The specific method is as follows:
[0156] 1. Synthesize cDNA
[0157] Use takara's PrimeScriptII 1st Strand cDNA synthesis Kit to synthesize cDNA according to the instructions:
[0158] system:
[0159] Volume (ul) Huh7.0 RNA 2 Primer P2 0.5 dNTP 4 RNase free ddH 2 o
6.5ul Total 13
[0160] 65°C for 5 minutes, follo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com