Design method of realizing Markov chain by using reversible single molecule reaction
A Markov chain and design method technology, applied in the field of chemical reaction network calculation, can solve problems such as not realizing Markov chain calculation, and achieve the effect of reducing the types and the number of reactions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
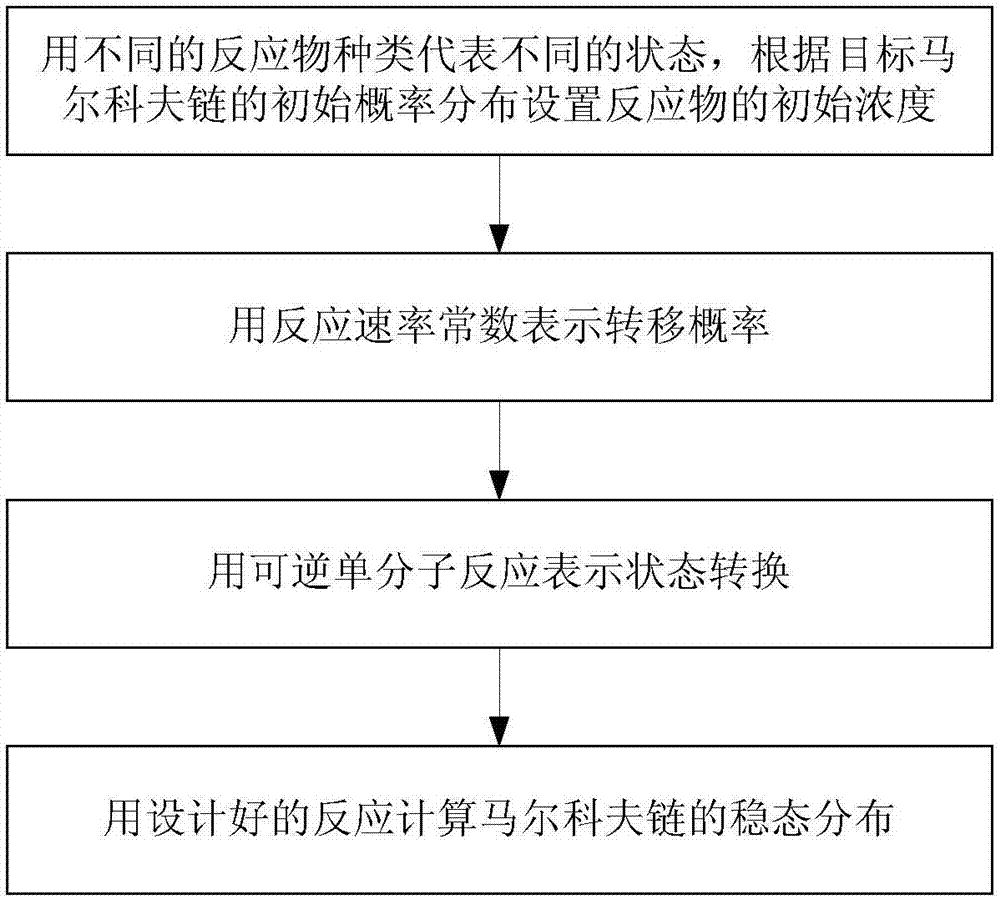
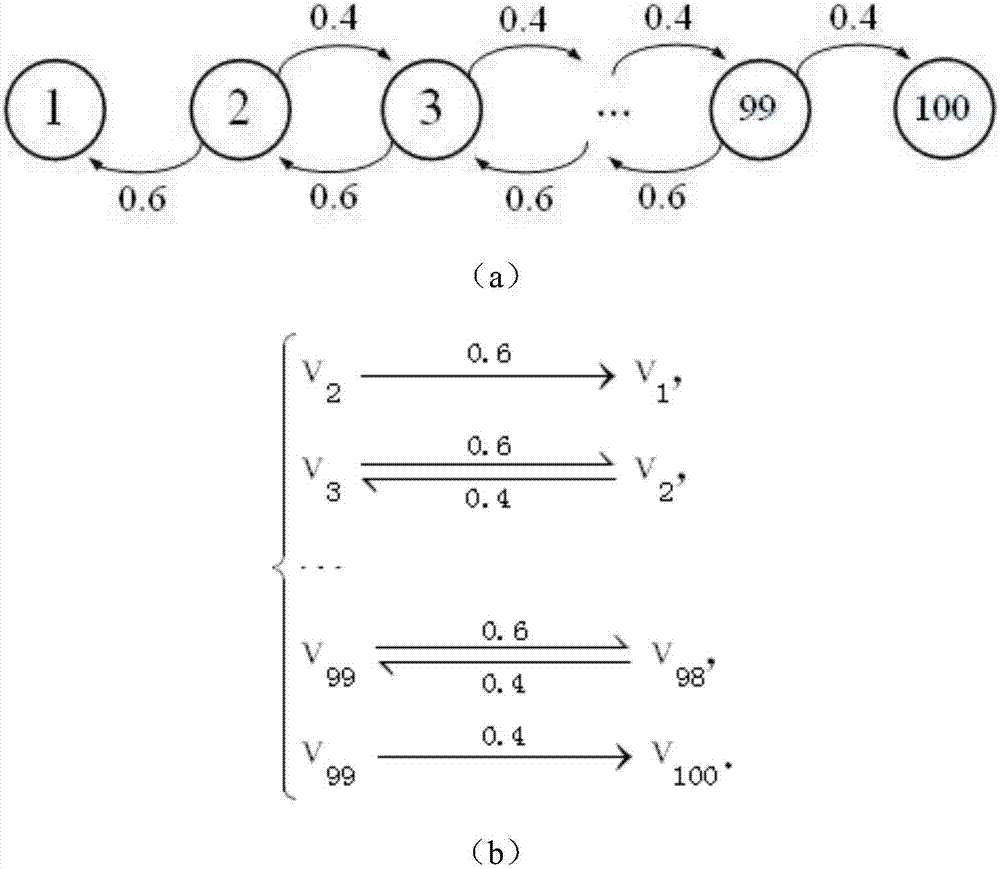
[0021] Such as figure 1 As shown, in the present invention, the design method of using a single molecule reversible reaction to realize a Markov chain includes the following steps:
[0022] (1) Use different reactant types ν i (i=1,2,...,k)ν i ,i=1,2,...k represents different states, and the initial concentration of reactants is set according to the initial probability distribution of the target Markov chain (the Markov chain to be solved for steady state distribution); that is, the initial concentration of all reactants Must be equal to or proportional to the initial state distribution of the target Markov chain;
[0023] (2) Use the reaction rate constant to express the transition probability k ij (i=1, 2,...,k j=1, 2,...,k), set the value of the reaction rate constant according to the transition probability;
[0024] (3) Use a reversible single molecule reaction Indicates state transition;
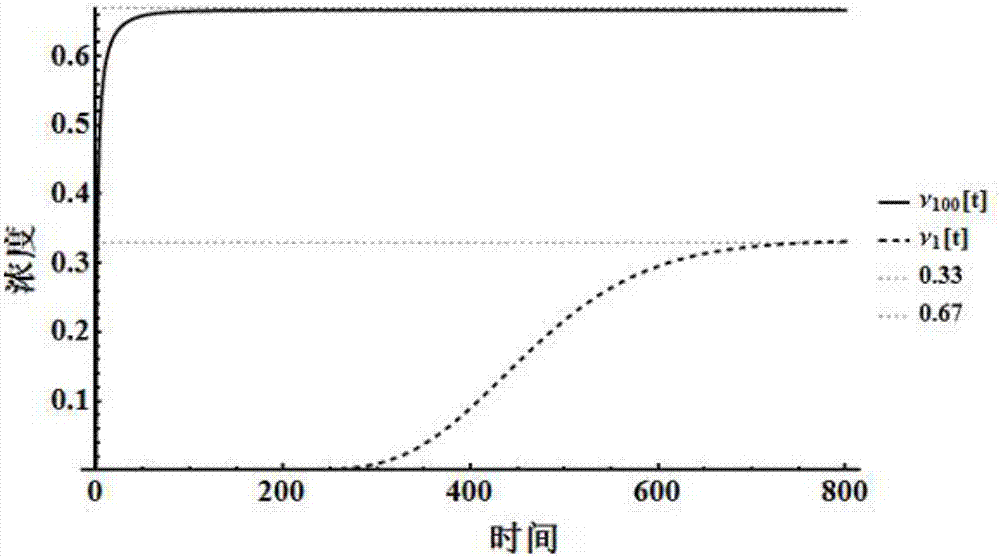
[0025] (4) Use the designed reaction to calculate the steady-state distribution of the Ma...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com