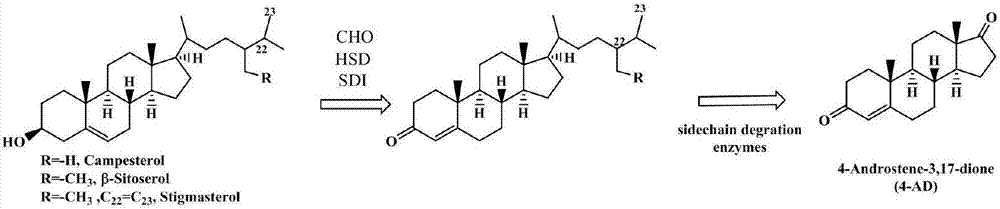
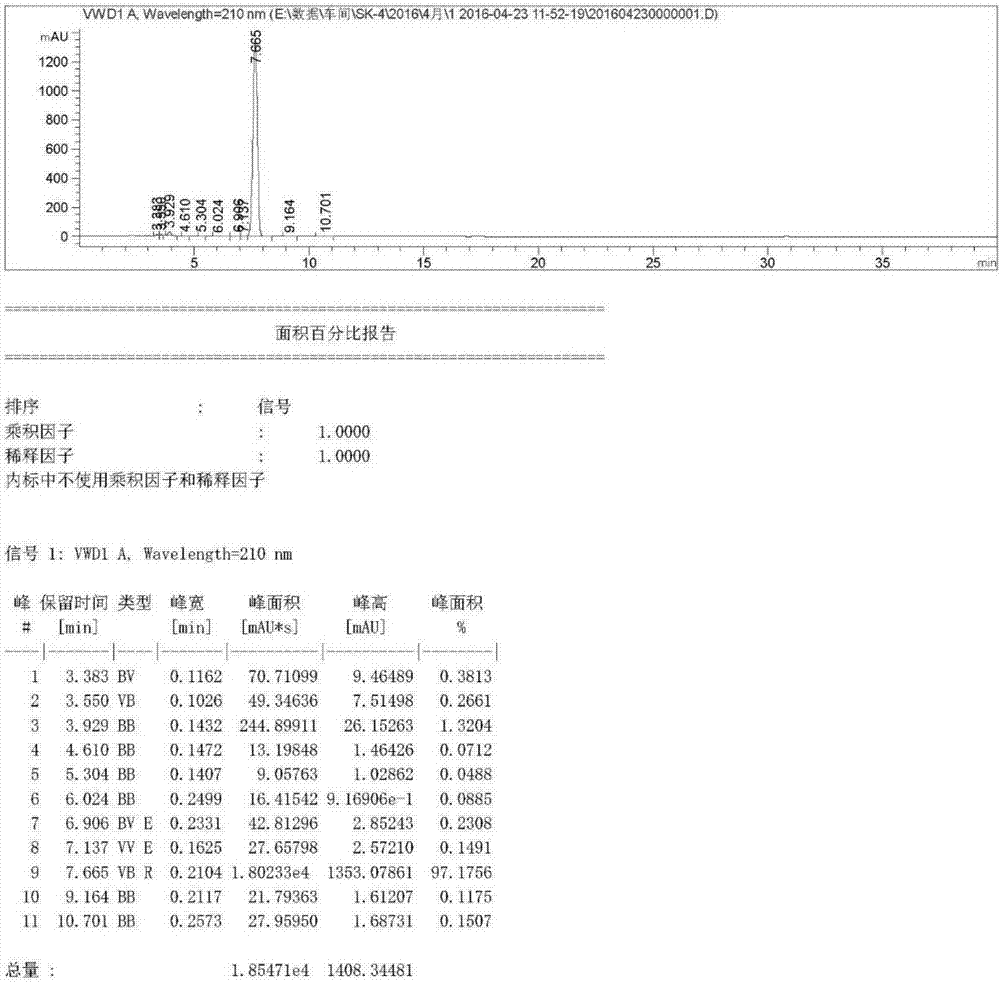
Defective mycobacterium and method for preparation of dehydroepiandrosterone from the same
A dehydroepiandrosterone and mycobacterium technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, bacteria, etc., can solve the problems of low efficiency of degrading phytosterol branched chains, no advantages, and low product purity. Achieve the effect of eliminating 3β-hydroxyl oxidation enzyme activity and reducing 3β-hydroxyl oxidation enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: Identification of phytosterol 3-hydroxy metabolizing enzyme gene of industrial production strain Mycobacterium GTF-69
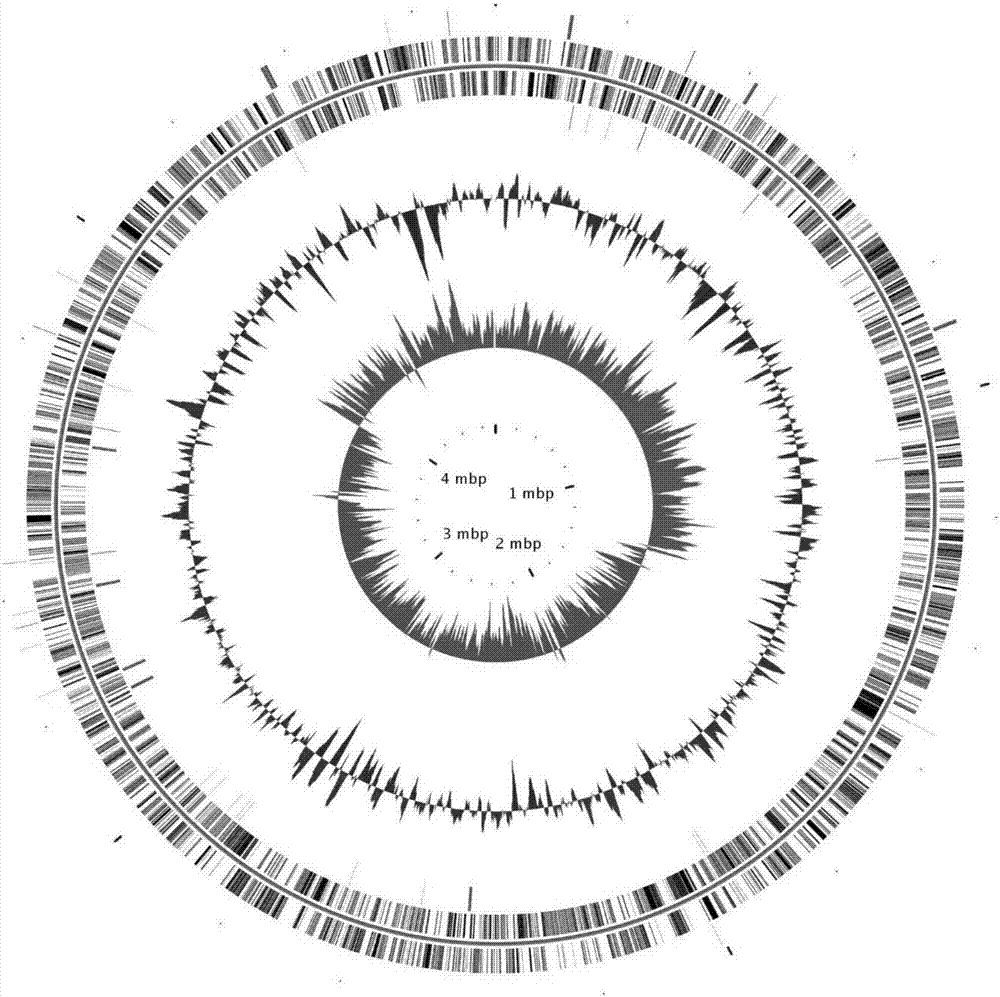
[0047] The Mycobacterium GTF-69 strain was analyzed by second-generation and third-generation high-throughput sequencing. Based on PacbioRSII data and IlluminaHiseq 4000 data, genome assembly software was used to analyze the number of chromosomes. The genome length was 5.421Mbp and the GC content was 1. 66.88%.
[0048] For the quality control analysis of the assembly results, IlluminaHiseq 4000 data is mainly used to evaluate the assembly results. The quality of single base analysis of the quality control result data is 0.9998, the quality of structural base analysis is 0.9920, the readability rate is 0.9842, and the repetition rate is 1.35 %.
[0049] After assembly, the GC bias of the strains was analyzed by GC-Depth. Draw its GC map according to the genome sequence, use the calculation method of (G–C) / (G+C) to perform GC skew analysis,...
Embodiment 2
[0051] Example 2: 3β-hydroxy metabolizing enzyme gene knockout
[0052] 1) Oligo DNA sequence design:
[0053] Use the MIT CRISPR Design (http: / / crispr.mit.edu / ) tool to design a pair of oligo DNA of about 20bp in the target DNA region. Select the appropriate Guide#1 sequence according to the score level, and design the sequence of two single-stranded oligos as follows (the part in lower case is the part to be complementary to the vector after digestion with Bsa I):
[0054] cho-F: caccGT AAG CAT GAA CAT GTA ACT CGC AAC
[0055] cho-R: aaacGT TGC GAG TTA CAT GTT CAT GCT TAC
[0056] In addition, a primer needs to be designed at the upstream and downstream of the site. This primer is used for subsequent PCR or sequencing to detect positive clones. This primer can amplify a DNA fragment of about 300 bp. The upstream primer is about 100 bp away from the mutation site, and the downstream primer is about 100 bp away from the mutation site Point about 200bp.
[0057] 2) Construc...
Embodiment 3
[0062] Embodiment 3: the preparation of mycobacterium competent cell
[0063] Add frozen glycerol bacteria to a test tube containing 2ml 7H9 medium to recover for 2-3 days. For bacteria that grow normally or are stored at 4 degrees, 1 to 5 ml can be added to a conical flask containing 50 mL of 7H9 (containing 0.1% Tween 80) liquid medium when inoculating. will concentration 10 8 About mycobacteria / ml were inoculated in conical flasks with 50 mL of 7H9 (containing 0.1% Tween-80) liquid medium with a 10% inoculation amount, and the bacteria were amplified by shaking at 37°C. At the same time, about 10 glass beads need to be added into the Erlenmeyer flask. Incubate at 37°C to OD 600nm = 0.4~0.5, add glycine with a final concentration of 1% to continue culturing, when OD 600nm When it reaches 0.8-1.4, start to prepare competent state.
[0064] Prepare 10% glycerol + 0.05% Tween 80 and filter through 0.2 μm filter, place in ice. Transfer 40ml of bacterial solution to a 50mL ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com