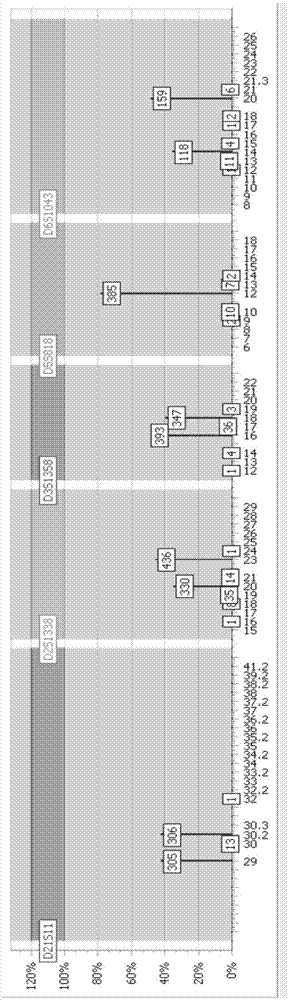
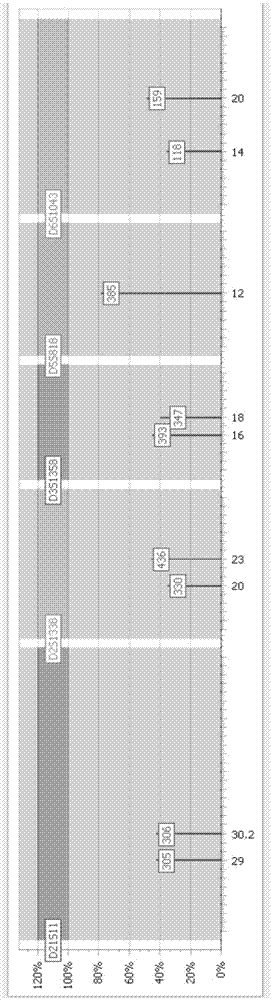
Processing method for high-throughput sequencing information of human short tandem repeat
A technology of tandem repeat sequence and processing method, which is applied in the field of high-throughput sequencing information processing of human short-fragment tandem repeat sequence, can solve the problem of inability to further detect minor differences in nucleic acid primary structure, difficulty in the number of analyzed loci, and limited resolution and other problems to achieve the effect of improving the detection rate and detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] Unless otherwise defined, all technical terms used in the embodiments of the present invention have the same meanings as commonly understood by those skilled in the art. In order to make the object, technical solution and advantages of the present invention clearer, the implementation manner of the present invention will be further described in detail below in conjunction with the accompanying drawings.
[0035] An embodiment of the present invention provides a method for processing high-throughput sequencing information of human short-fragment tandem repeat sequences, the processing method comprising the following steps:
[0036] Step 101: Obtain the STR high-throughput sequencing information of a single chip as the original sequence, and filter the original sequence according to the preset sequencing length, retain the sequence with the preset sequencing length, and form the first sequence to be processed.
[0037] It can be understood that the preset sequencing length ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com