A method for analyzing the genetic diversity of Tsaoguo germplasm resources based on srap markers
A technology of genetic diversity and fruit, applied in the field of genetic diversity analysis of grass and fruit germplasm resources based on SRAP markers, which can solve the problem of large experimental error, limitation of identification of grass and fruit resources and genetic diversity analysis, and morphological characters susceptible to the environment. Condition effects and other problems, to achieve the effect of clear bands, accelerated identification speed, and stable and reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Method for analyzing genetic diversity of Tsaoguo germplasm resources based on SRAP markers
[0046] (1) Tsaoguo genomic DNA was extracted by 2*CTAB method for future use.
[0047] (2) Caoguo SRAP-PCR reaction system optimization, the optimized reaction system is characterized in that: 25 μ L reaction system includes: 10 × PCR buffer (without Mg 2+ )3.0μL, Taq enzyme 1U, Mg 2+ 1.5mmol / L, dNTP 0.25mmol / L, primer 0.2μmol / L and template DNA 30ng.
[0048] (3) Improvement of the PCR amplification reaction program, the improved program: pre-denaturation at 95°C for 5 minutes, denaturation at 95°C for 1 minute, annealing at 35°C for 1 minute, extension at 72°C for 1 minute, 35 cycles, final extension at 72°C for 10 minutes, and storage at 4°C .
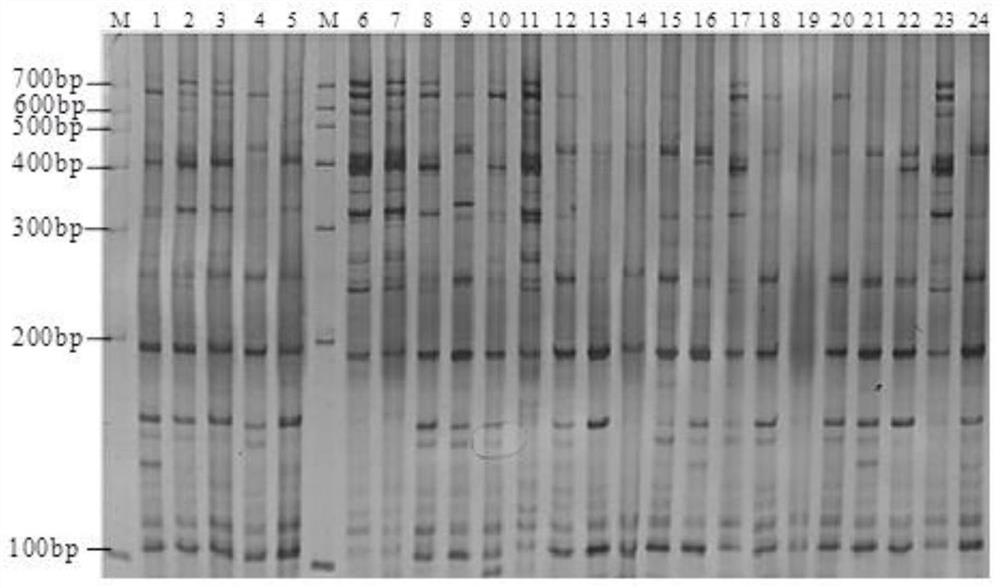
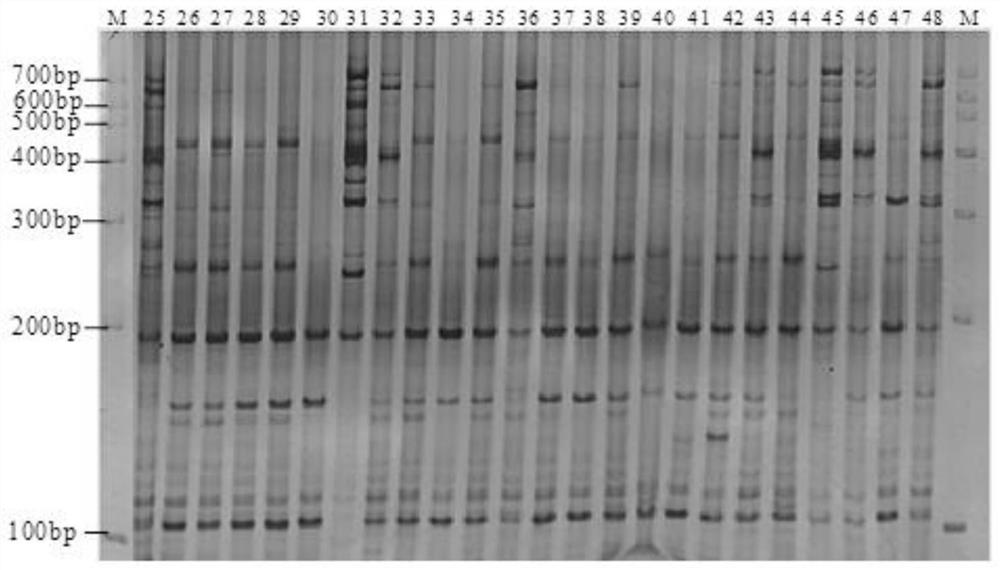
[0049] (4) Screening of SRAP polymorphic primers. From 153 pairs of SRAP primers, 12 pairs of stable, clear and reproducible polymorphic primers were screened out for PCR amplification reaction of different populations o...
Embodiment 2
[0051] The extraction of embodiment 2DNA
[0052] (1) Experimental materials
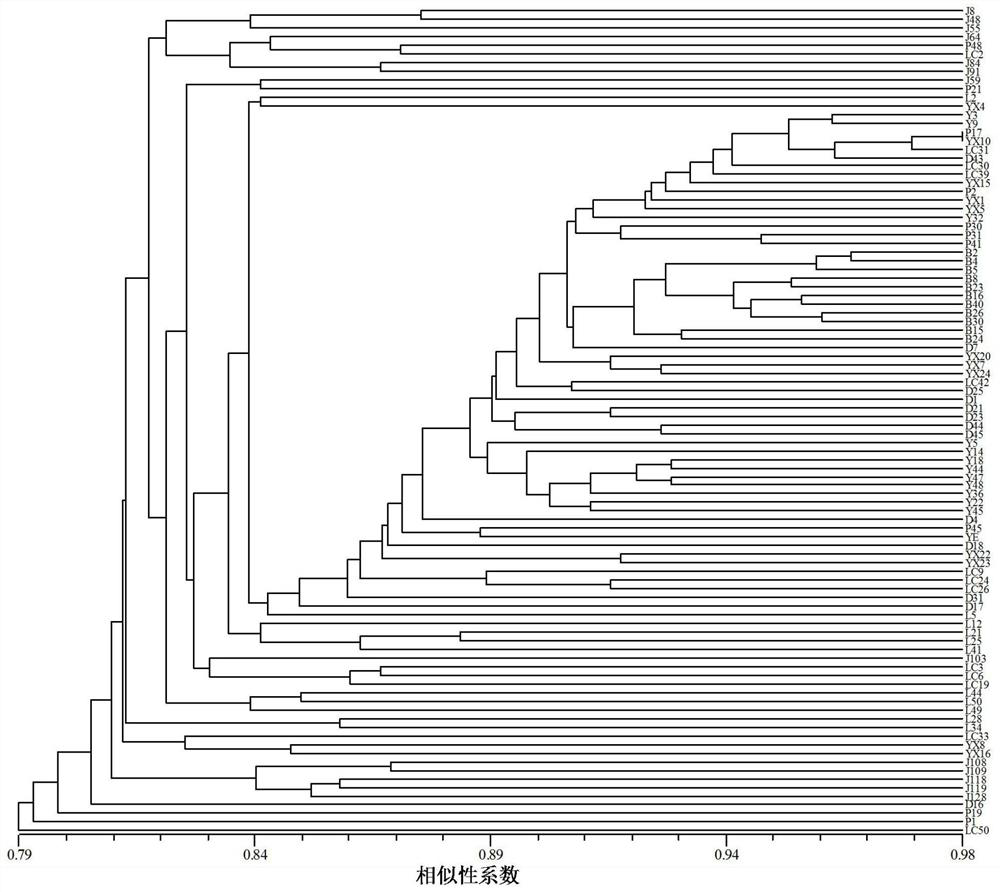
[0053] The 96 Tsaoguo used in this experiment were collected from 8 Tsaoguo producing areas in Yunnan Province, including 13 from Jinping County, 12 from Yuanyang County, 11 from Luchun County, 10 from Pingbian County, 14 from Pu’er Lancang County, Baoshan There are 11 copies in the city, 13 copies in Dehong Lianghe County, and 12 copies in Lincang Yun County. See Table 1 and Table 2 for details.
[0054] Table 1 Test material number (1-48) and sampling points
[0055]
[0056]
[0057] Table 2 Test material number (49-96) and sampling points
[0058]
[0059]
[0060] (2) Extraction of DNA
[0061] Young leaves were collected at the same time as the germplasm resource investigation for genome-wide DNA extraction. DNA was extracted by 2*CTAB method, and the extracted DNA samples were dissolved in TE buffer and stored at -20°C for later use. Before amplification, dilute the DNA sampl...
Embodiment 3
[0062] Embodiment 3PCR reaction system optimization and primer screening
[0063] Using the orthogonal design method, the Mg 2+ , dNTPs and primer concentration, TaqDNA polymerase and the amount of template DNA were screened, and the SRAP-labeled PCR reaction system suitable for Tsaoguo was obtained. The orthogonal design of SRAP-PCR is shown in Table 3. After three repeated tests of primers me1em15, me4em8, me7em1, and me8em14, 10×PCR buffer (excluding Mg 2+ )3.0μL, Taq enzyme 1U, Mg 2+ 1.5mmol / L, dNTP 0.25mmol / L, primer 0.2μmol / L and template DNA 30ng comprehensive amplification effect is the best.
[0064] Table 3 SRAP-PCR reaction L16 (4 5 ) Orthogonal Experiment Design Table
[0065]
[0066]
[0067] The PCR amplification reaction program of the above reaction system is an improved program: pre-denaturation at 95°C for 5 minutes, denaturation at 95°C for 1 minute, annealing at 35°C for 1 minute, extension at 72°C for 1 minute, 35 cycles, and finally extension ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com