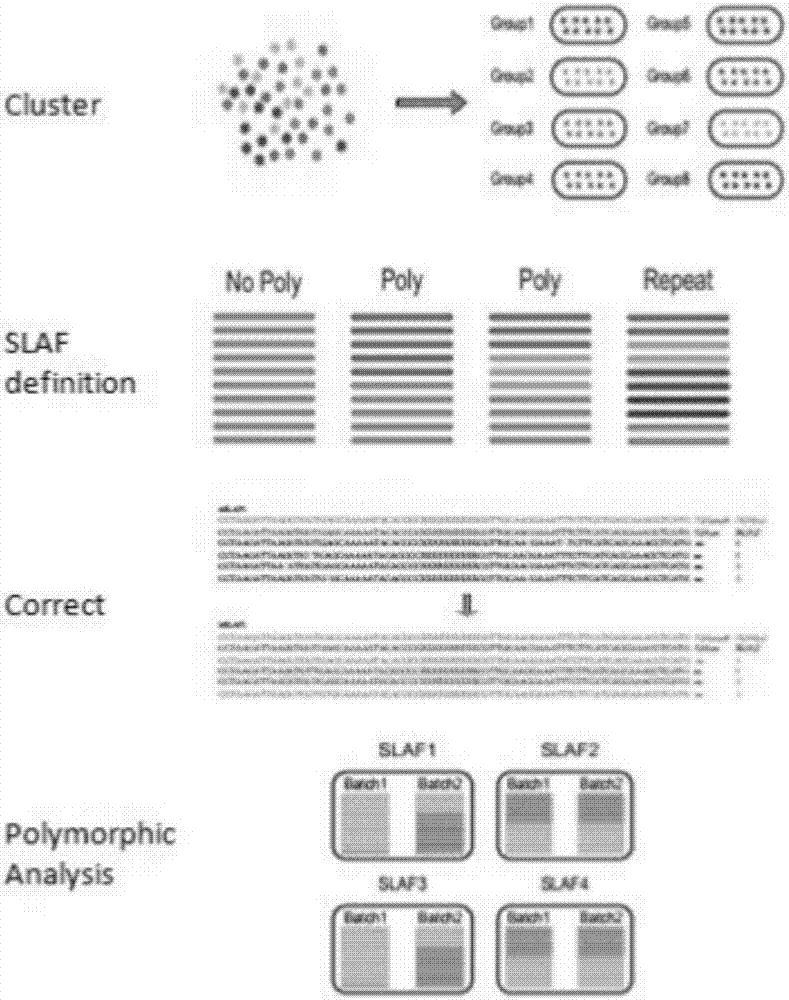
Method for identifying genetic relationship by developing SNP molecular marker of miscanthus with SLAF-seq technology
A phylogenetic relationship, slaf-seq technology, applied in the field of identification of phylogenetic relationship and phylogenetic evolution research of Miscanthus plants, to achieve the effect of reducing genome complexity, reducing complexity and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] Select 19 offspring samples of Miscanthus whose female parent is known to be Nandi, and samples of Miscanthus, Digi, and Nandi that met the female parent in flowering period were selected as 30 suspected male parents and 1 female parent, a total of 50 materials.
[0029] 1 Reference Genome Determination
[0030] According to the genome size and GC content of Miscanthus plants, the sorghum genome was finally selected as the reference genome for enzyme digestion prediction.
[0031] The specific information is as follows:
[0032] a) Sequencing species information: Miscanthus, the actual genome size is about 2.65Gb, and the GC content is 41.39%;
[0033] b) Reference species information: Sorghum bicolor genome, the assembled genome size is 738.54Mb, GC content is 43.90%, download address: http: / / phytozome.jgi.doe.gov / pz / portal.html#! info? alias = Org_Sbicolor.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com