A method for gene association analysis based on deep learning algorithm
A technology of deep learning and association analysis, applied in the field of bioinformatics, can solve problems such as inability to handle variable-length sequences, achieve high accuracy, save time, and reduce burden
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
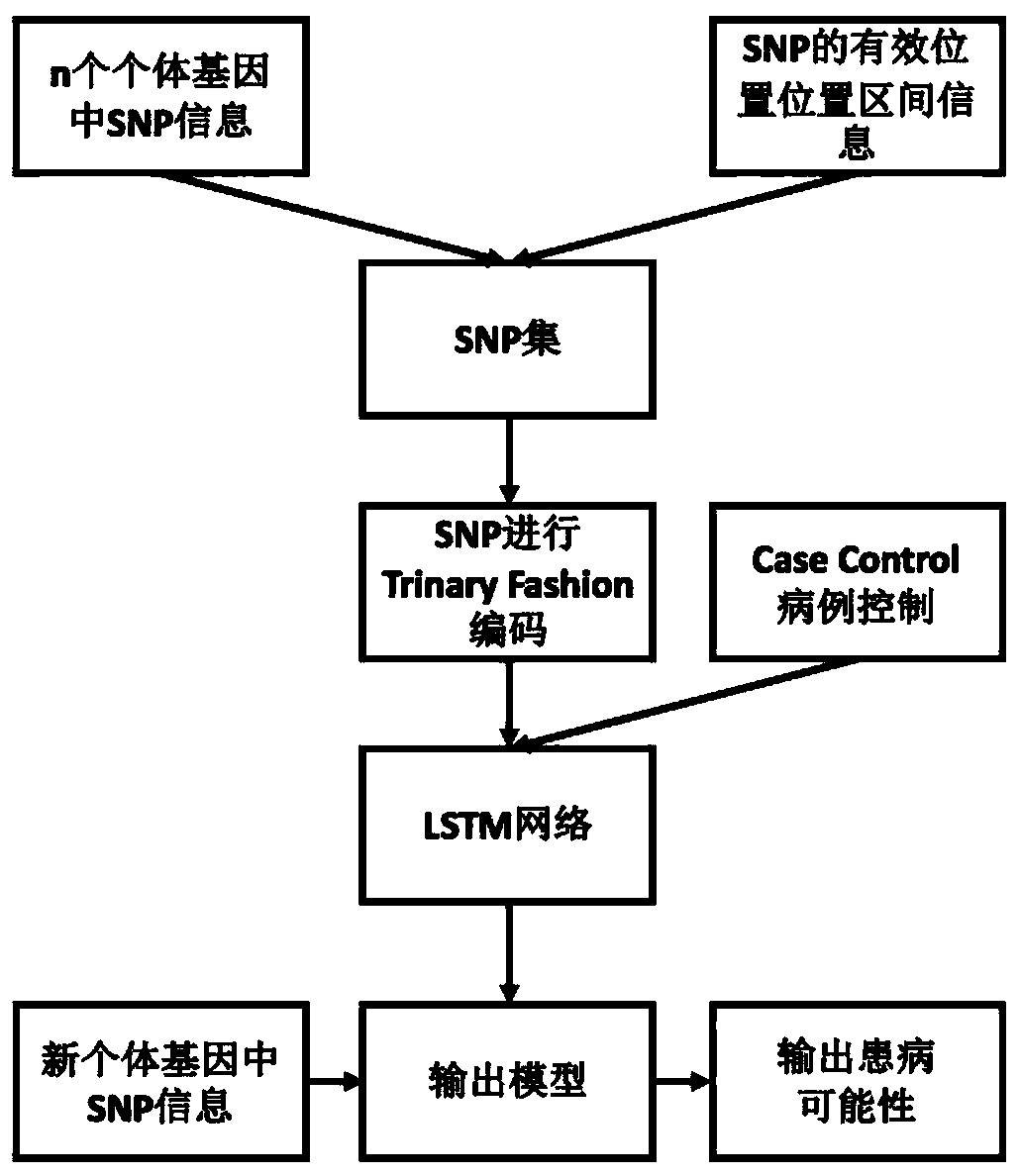
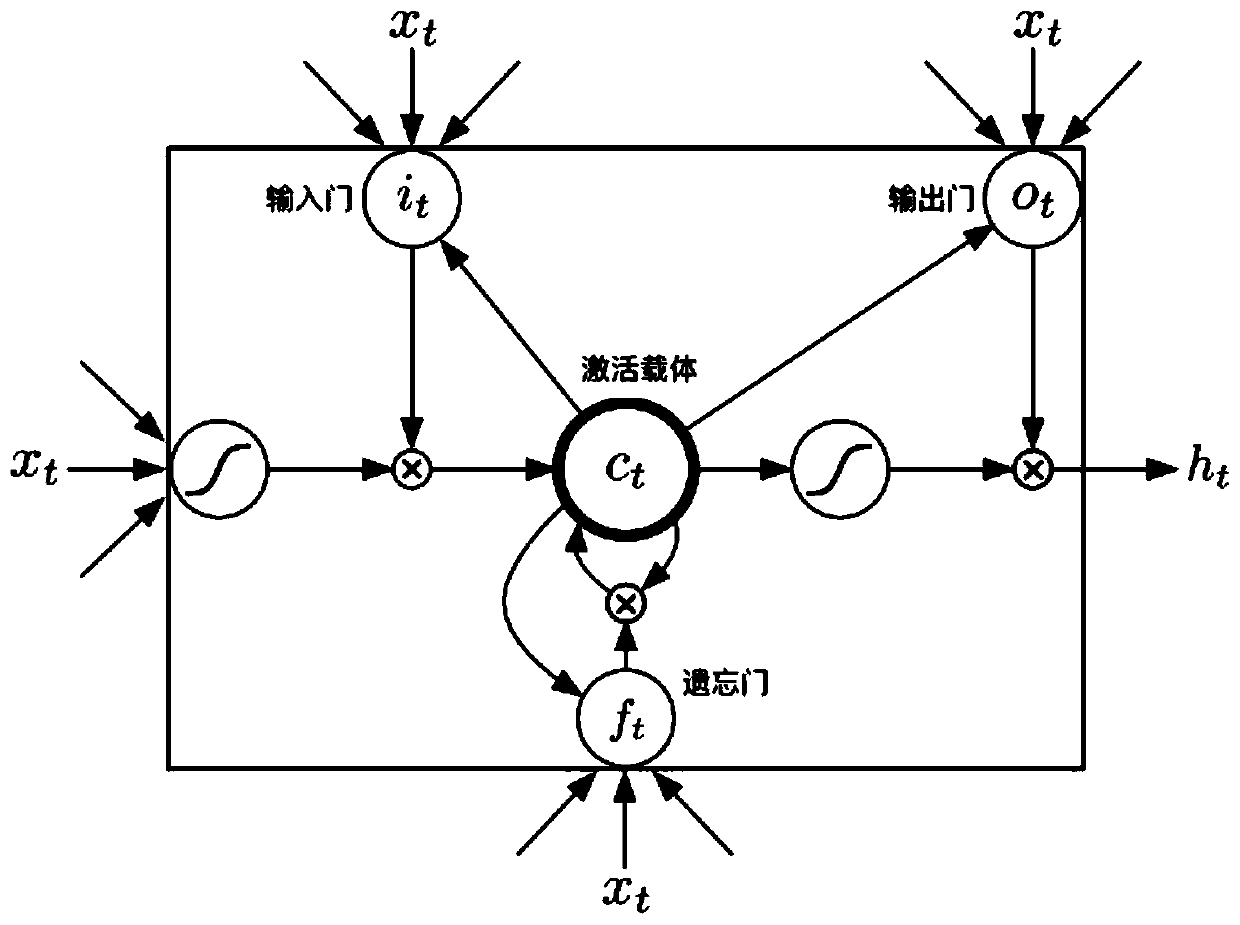
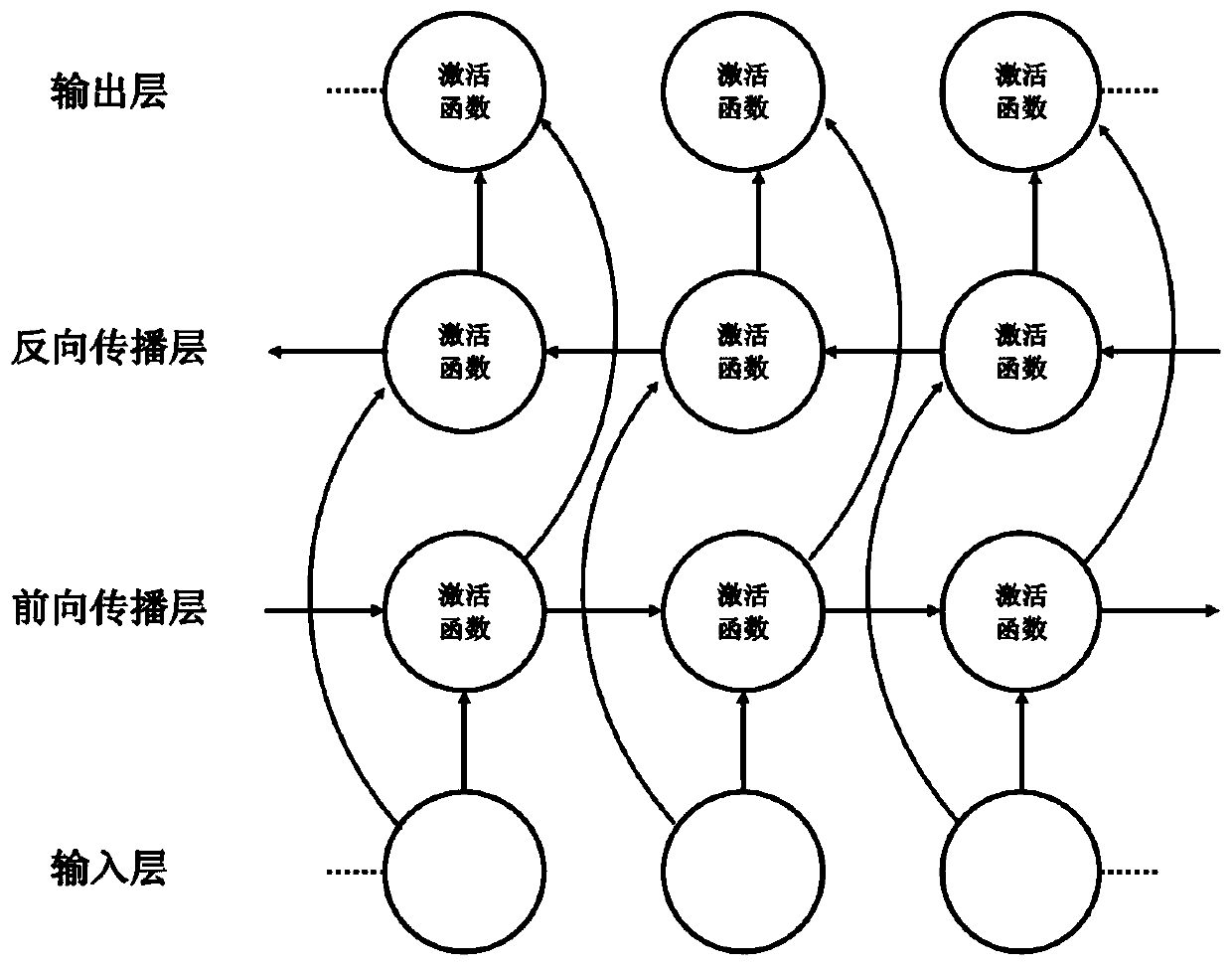
[0036] Reference manual attached figure 1 In the following, the technical solutions of the present invention will be specifically described through implementation, but the present invention is not limited to the following embodiments.
[0037] Step 1: According to the existing biological knowledge, segmentation is performed at the chromosome level according to the gene distribution, and the effective position interval information of the SNP is obtained according to the location of the gene, for subsequent segmentation of the SNP. The sample genes of CEU (NorthernEuropeans from Utah) are used as a simulation.
[0038] Step 2: Suppose that based on the case control of the population, the gene sequence of n independent individuals is used to translate the SNP at the chromosome level to obtain the required input data.
[0039] Step 3: As attached Figure 4 As shown, according to the position information obtained in step 1, the SNP sequence obtained in step 2 is grouped according to the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com