RPA (Recombinase Polymerase Amplification) specific primers and kit for detecting vibrio parahaemolyticus in food and application
A specific technology for Vibrio hemolyticus, applied in the direction of DNA / RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of different temperatures and limited application range, etc., to achieve sensitive detection methods and a wide range of applications , the effect of short detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] The selection of embodiment 1 target gene and the design of RPA primer
[0044] The known specific genes of Vibrio parahaemolyticus were analyzed by bioinformatics, and the target gene toxR was selected for detection. Through the public BLAST software in Genbank, the sequence of the toxR gene was compared with other microorganisms, and the sequence segment with higher specificity was selected (GeneBank ID: AB029908). Use the software Primer 5.0 to design specific primers in this sequence according to the requirements of RPA primer design, and use bioedit to compare the primers with species with high homology of Vibrio parahaemolyticus, see figure 1 , 2 , from which primer pairs with low homology to other species were selected. The invention designs RPA primers of Vibrio parahaemolyticus, and the product size is 426bp. The specific sequence is as follows:
[0045] Upstream primer F14: 5'-ACTCGTATGAGAACGTGACATTGCGTATTT-3'; (SEQ ID No: 1)
[0046] Downstream primer R1...
Embodiment 2
[0047] The design and construction of embodiment 2 amplification internal standard
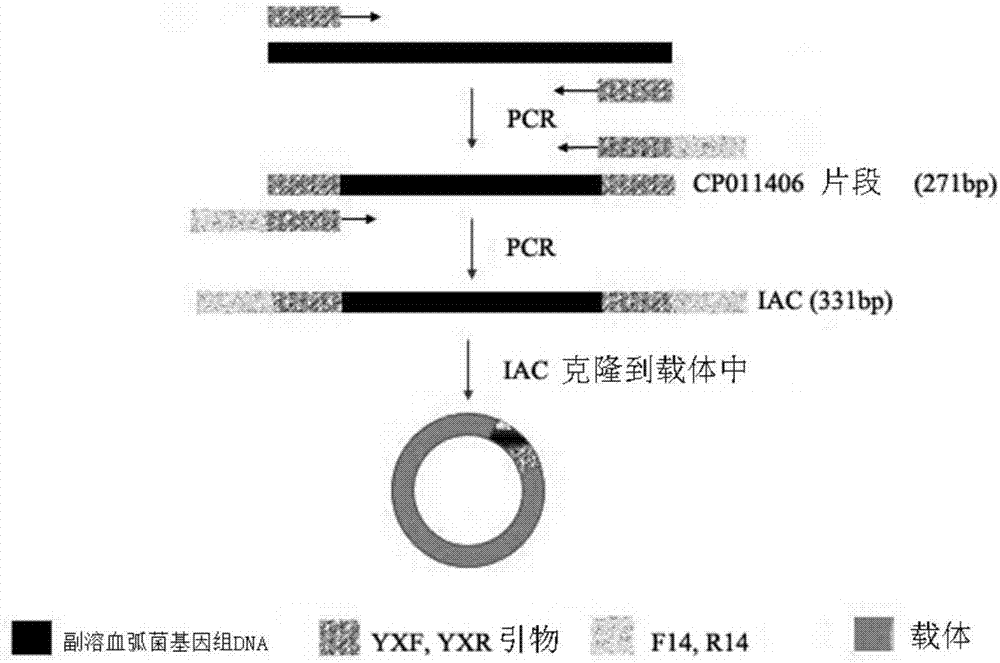
[0048] After amplifying the internal standard primers YXF and YXR, and PCR amplification, the reaction conditions were: 94°C for 5 min; 94°C for 30 s, 59°C for 60 s, 72°C for 30 s, 30 cycles; 72°C for 10 min. The Vibrio parahaemolyticus fragment CP011406 (271bp) with a very low degree of homology to the target gene fragment was obtained. Target detection primers F14 and R14 were connected to the 5' ends of the amplification internal standard primers YXF and YXR to obtain long primers IACF and IACR. CP011406 was amplified by long primers IACF and IACR to obtain a 331bp amplified internal standard fragment. The reaction conditions were: 94°C for 5min; 94°C for 30s, 57°C for 30s, 72°C for 60s, 30 cycles; 72°C for 10min. The designed amplified internal standard sequence was artificially synthesized and cloned into the vector Mach1-T1 (Beijing Quanshijin Co., Ltd., pEASY-T1 Cloning Kit, item numbe...
Embodiment 3
[0056] Example 3. Preliminary construction of the RPA-IAC detection method for detecting Vibrio parahaemolyticus
[0057] 1. Extraction of DNA from the sample to be tested
[0058] Take 1mL of the bacterial solution in a 1.5mL centrifuge tube, centrifuge at 12000rpm for 1min, discard the supernatant, and extract the bacteria of Vibrio parahaemolyticus according to the kit operation (Bacterial Genomic DNA Extraction Kit, Tiangen Biotechnology Co., Ltd., Cat. No.: DP302). Liquid DNA. The obtained DNA was stored at -20°C for future use.
[0059] 2. RPA-IAC amplification
[0060] Using TwistAmp Basic kit (TwistDX, TABAS03KIT, Cambridge, UK), the configuration method of RPA-IAC for detecting Vibrio parahaemolyticus is as follows:
[0061] Add the following solutions to the 0.2 mL TwistAmp reaction tube containing lyophilized enzyme powder:
[0062]
[0063] Vortex the above mixture to mix well, and centrifuge briefly to remove water beads on the wall. Add 280mM MgAc into a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com