Primer combination for simultaneously identifying Chinese herb ginseng, American ginseng and radix notoginseng and application of primer combination
A combination of primers and American ginseng technology, which is applied in the direction of recombinant DNA technology, biochemical equipment and methods, and microbial measurement/inspection, can solve the problems of drug safety damage, mixed use of adulterants, and inability to identify adulterants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0103] Embodiment 1, primer design
[0104] Extensive sequence analysis of the genomes of various Panax genus species yielded several primers for the identification of Panax ginseng, Panax notoginseng and American ginseng. Each primer was pre-tested to compare the performance of sensitivity and specificity, and finally three sets of primer pairs for identifying ginseng, Panax notoginseng and American ginseng were obtained.
[0105] The primer pair (primer pair I) used to identify ginseng consists of primer F1 and primer F2 (5'→3'):
[0106] F1 (sequence 1 of the sequence listing): TAACAATACCGGGCTGATAC;
[0107] F2 (sequence 2 of the sequence listing): CCAGTTAAGGACAGGAG;
[0108] The primer pair (primer pair II) used to identify Panax notoginseng consists of primer F3 and primer F4 (5'→3'):
[0109] F3 (sequence 3 of the sequence listing): AGGGATGAGGGGTGCGTAG;
[0110] F4 (sequence 4 of the sequence listing): CGACATGAGAAGAGGGCTTTTA;
[0111] The primer pair (primer pair II...
Embodiment 2
[0115] Embodiment 2, identification method optimization
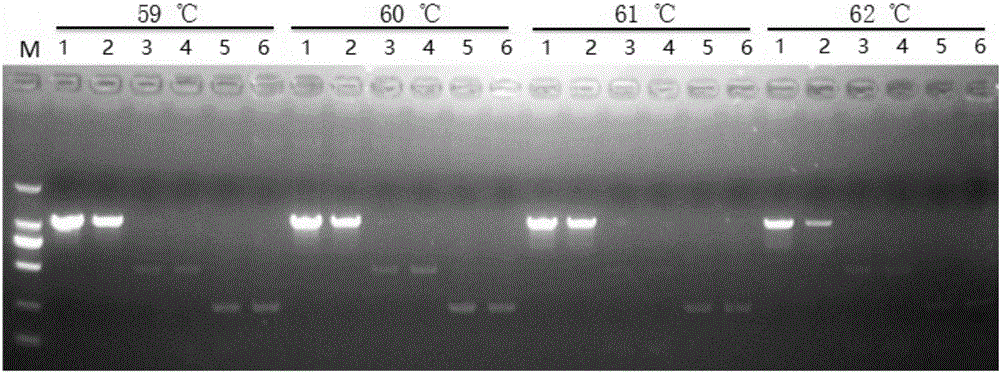
[0116] 1. Optimization of annealing temperature
[0117] The samples to be tested are: ginseng (serial numbers 1 and 2 in Table 1), Panax notoginseng (serial numbers 10 and 11 in Table 1), American ginseng (serial numbers 16 and 17 in Table 1).
[0118] 1. Extract the genomic DNA of the sample to be tested.
[0119] 2. Take the genomic DNA obtained in step 1 as a template, and use the primer combination prepared in Example 1 to carry out PCR amplification.
[0120] PCR amplification system (25 μL): 2.5 μL 10×buffer, 1.6 μL dNTP (2.5 mM), 0.4 μL primer F1, 0.4 μL primer F2, 0.4 μL primer F3, 0.4 μL primer F4, 0.4 μL primer F5, 0.4 μL μL primer F6, 0.2 μL SpeedStar HS Taq DNA polymerase, 2 μL template (DNA content is 10 ng), 16.3 μL sterile double distilled water.
[0121] PCR reaction program: pre-denaturation at 95°C for 5 min; denaturation at 95°C for 10 s, annealing for 10 s, extension at 72°C for 20 s, a total of ...
Embodiment 3
[0140] Embodiment 3, establishment of identification method
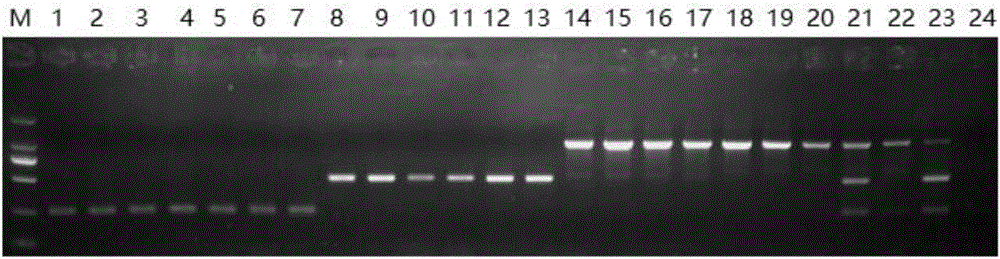
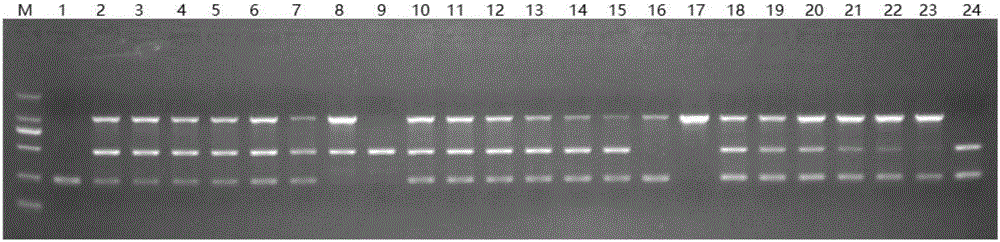
[0141] 1. Agarose gel electrophoresis
[0142] 1. Extract the genomic DNA of the sample to be tested.
[0143] 2. Using the genomic DNA obtained in step 1 as a template, the primer combination prepared in Example 1 was used for PCR amplification to obtain an amplification product.
[0144] PCR amplification system (25 μL): 2.5 μL 10×buffer, 1.6 μL dNTP (2.5 mM), 0.3 μL primer F1, 0.3 μL primer F2, 0.2 μL primer F3, 0.2 μL primer F4, 0.25 μL primer F5, 0.25 μL primer F6, 0.2 μL SpeedStar HS Taq DNA polymerase, 2 μL template DNA, 17.2 μL sterile double distilled water.
[0145] PCR reaction program: pre-denaturation at 95°C for 5 min; denaturation at 95°C for 10 s, annealing at 60°C for 10 s, extension at 72°C for 20 s, a total of 35 cycles; extension at 72°C for 3 min.
[0146] 3. Perform 1.5% agarose gel electrophoresis on the amplified product obtained in step 2, and judge as follows according to the result:
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com