Tomato 212 SNP loci as well as applications thereof to identification of variety authenticity and seed purity of Lycopersicon esculentum
A technology of authenticity and variety, applied in the biological field, it can solve the problems of low polymorphism, small genetic variation of tomato, and few types of enzymes.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Embodiment 1, the preparation of the primer set that is used to detect 212 SNP site genotypes
[0074] (1) Obtain information on 7,000 SNP sites covering 12 tomato chromosomes by searching literature (Sim SC, et al. (2012) High-Density SNP Genotyping of Tomato (Solanum lycopersicum L.) Reveals Patterns of Genetic Variation Due to Breeding . Plos One 7(9): e45520-e45520.).
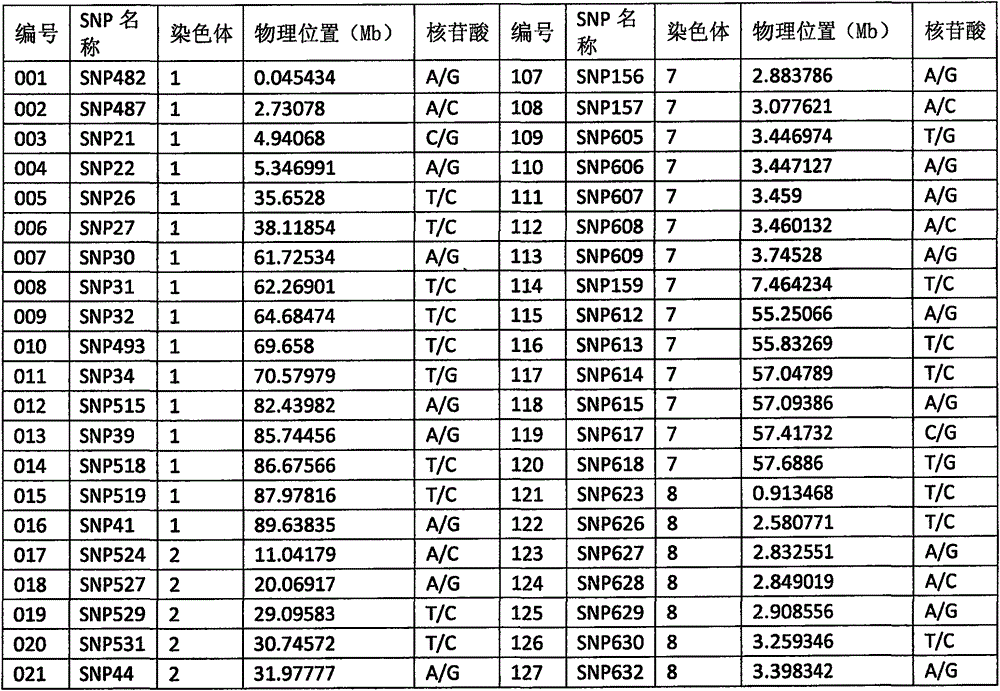
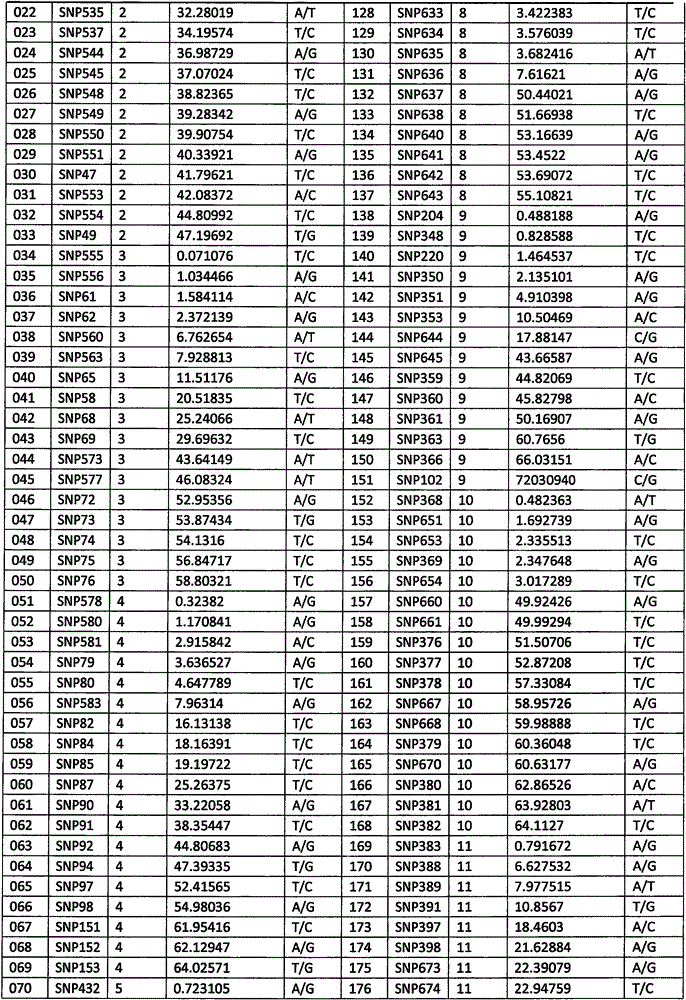
[0075] (2) 212 SNP sites were selected from the above SNP sites, and the physical positions and flanking sequences of these 212 SNP sites were determined based on the whole genome sequence (version number SL2.50) of tomato variety Heinz 1706 Yes, the 212 SNP sites are numbered 001-212, and the 212 SNP sites are specifically shown in Table 1.
[0076] Table 1, 212 SNP site information
[0077]
[0078]
[0079]
[0080]Three primers are required to detect the deoxynucleotide of each SNP site, two upstream primers (denoted as P1 and P2) and one downstream primer (denoted as P3). The detecti...
Embodiment 2
[0082] Example 2. Using a set of primers for detecting the genotypes of 212 SNP sites to identify the authenticity of tomato varieties
[0083] (1) The tomato material for constructing the map was provided by Mr. Li Changbao from the Vegetable Center of Beijing Academy of Agriculture and Forestry Sciences. From 402 materials, 100 varieties were randomly selected (Table 2).
[0084] Table 2, 100 test tomato varieties
[0085]
[0086]
[0087] (2) Extract the DNA of the tested tomato varieties
[0088] Using the CTAB method to extract the DNA of the tested variety: take 20mg-30mg of young tomato leaves into a 2.0mL centrifuge tube, grind in a grinder for 10min; add 750μL of preheated CTAB extract, shake well, and put in a water bath at 65°C for 45min; add 750 μL of chloroform, mix up and down for 3 minutes, centrifuge at 12,000 g for 5 min; pipette 500 μL of supernatant into another 2.0 mL centrifuge tube, add 0.7 times the volume of pre-cooled isopropanol, invert the ce...
example 1:Allele1/Allele1,A/C,Allele1A,A/A。 example 2
[0099] According to the data read by the instrument's own software, the data obtained by the above two detection platforms are analyzed to form the fingerprint of each tomato variety. The genotype data of the homozygous site is recorded as Allele1 / Allele1 or Allele2 / Allele2, and the genotype data of the heterozygous site is recorded as Allele1 / Allele2, where Allele1 and Allele2 are the two alleles on the mutation site; The genotype data of missing loci were recorded as 0. Example 1: The genotype of a sample at a certain site is Allele1 / Allele1, the allele at this site is A / C, Allele1 represents base A, and the genotype at this site is recorded as A / A. Example 2: The genotype of the sample at a certain site is Allele1 / Allele2, the allele at this site is A / C, Allele1 represents base A, Allele2 represents base C, and the genotype at this site is recorded for A / C.
[0100] Determine whether the tomato to be tested is the same variety according to the test results: when the numbe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com