Method for accurately obtaining exosome miRNA (micro Ribonucleic Acid)
An exosome and precise technology, applied in the field of RNA detection, can solve the problems affecting the detection sensitivity and accuracy, the accuracy and purity of miRNA, etc., to achieve comprehensive and accurate expression profiles, avoid bias, and achieve the effect of accurate counting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
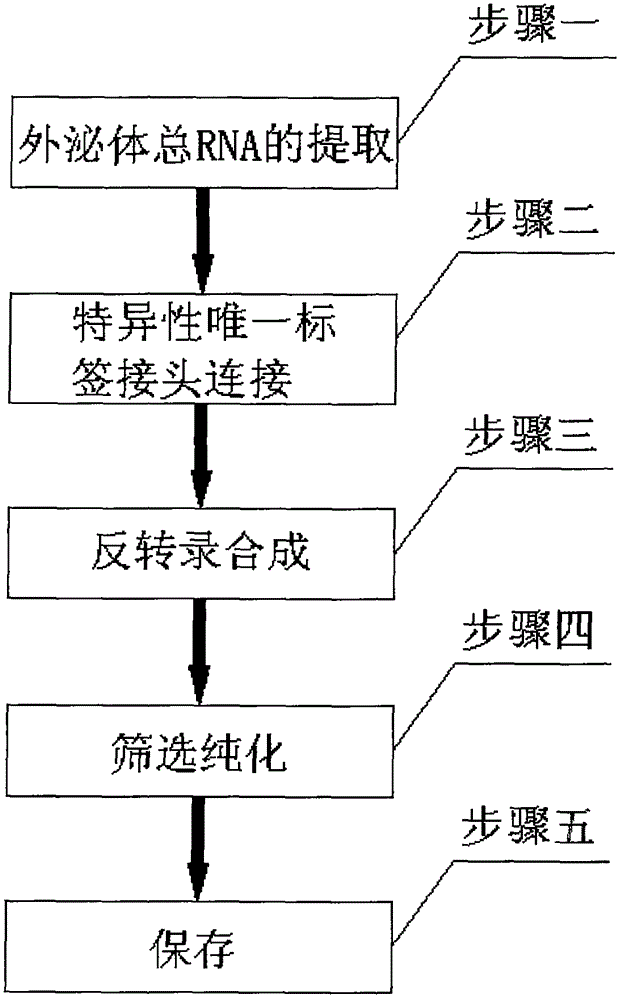
[0031] Draw 10ml of peripheral blood from the subject into an EDTA anticoagulant tube, gently invert it up and down 7 times and mix thoroughly, and perform the following treatment within 5 hours on the day of blood collection: Use the Wako exosome extraction kit to obtain exosomes from human peripheral blood.
[0032] After all the exosomes were isolated, the total RNA in the exosomes was extracted. Specifically, the Tiangen RNAprep pure blood total RNA extraction kit was used. After the total RNA was extracted, the total RNA was dissolved back into 14 μL of RNase-freewater, and then Preliminary quantification was performed based on Qubit2.0 (Invitrogen, QubitRNAHSAssayKits).
[0033] Connect the specific unique label linker to the exosome total RNA to obtain the total RNA with the specific linker; the specific unique label linker is:
[0034] 5′-rAppCTGTAGGCACCATCAATATTGCGCACGA-NH 2 3′
[0035] Mix the total RNA with specific adapters and other reverse transcription materia...
Embodiment 2
[0037] Put 10ml of human body fluid into an EDTA anticoagulant tube, invert up and down 6 times and mix thoroughly, and perform the following treatment within 4 hours on the day of blood collection: Use Wako exosome extraction kit to obtain exosomes.
[0038] After all the exosomes were isolated, the total RNA in the exosomes was extracted. Specifically, a total RNA extraction kit was used. After the total RNA was extracted, the total RNA was dissolved back into 10 μL of RNase-freewater, and then based on Qubit2.0 ( Invitrogen, QubitRNAHSAssayKits) for preliminary quantification.
[0039] Connect the specific unique label linker to the exosome total RNA to obtain the total RNA with the specific linker; the specific unique label linker is:
[0040] 5′-rAppCTGTAGGCACCATCAATGAACTCGTCGA-NH 2 3′
[0041] Mix the total RNA with specific adapters and other reverse transcription materials, mix the solution evenly, place it on a PCR machine, and incubate at 43°C for 55 minutes to rev...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com