A method for analyzing the gene sequence structure of non-model organism transcriptome
A gene sequence and structure analysis technology, applied in the field of gene analysis bioinformatics, can solve the problems of high false positives and inability to guarantee the function of protein sequences, etc., and achieve high reliability results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
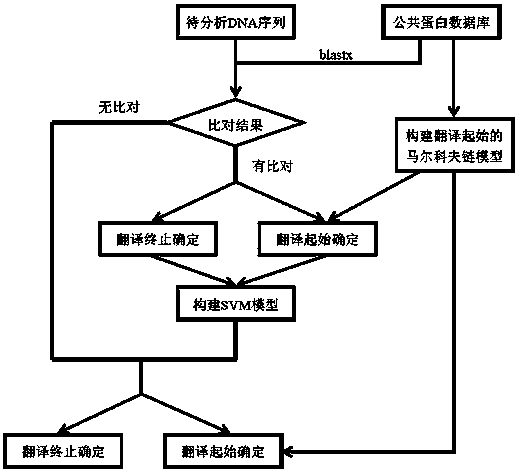
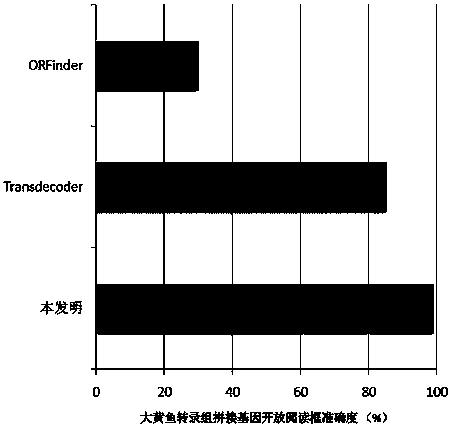
[0029] see Figure 1~2 In this example, a method for analyzing the gene sequence structure of the transcriptome of a non-model organism provided by the invention is used to analyze the gene sequence structure of the transcriptome splicing product of large yellow croaker.
[0030] Large yellow croaker (Larimichthys crocea), commonly known as yellow croaker and yellow croaker, belongs to the genus Yellow croaker of the family Perciformes of the order Perciformes and is an important economic fish in the coastal waters of my country. At present, large yellow croaker is one of the seawater fish with the largest amount of nursery and breeding in my country. The annual output has exceeded 120,000 tons, and the annual direct economic output value is billions of yuan. A comprehensive analysis of the gene sequence of large yellow croaker is an important genetic resource for the genetic research of large yellow croaker, and it is the basis for exploring the genetic genes of important eco...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com