Aptamer for osteosarcoma detection and kit for osteosarcoma detection
A kit, a technology for bone cancer, applied in the directions of measuring devices, biochemical equipment and methods, instruments, etc., can solve the problems of limited detection concentration and improved accuracy, and achieve the effects of simple method, time saving, and rapid separation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
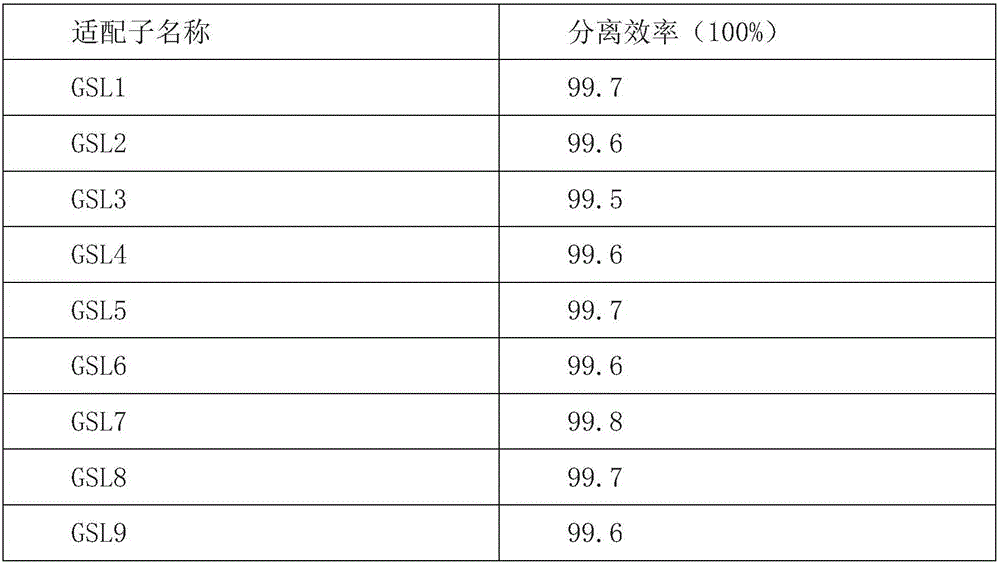
[0072] Embodiment 1: nucleic acid aptamer screening
[0073] From a random oligo DNA library synthesized in vitro, 5'-ATTCGACATGCCTGGACATA(N36)CATGCCACATGACCAAGGCA-3'.
[0074] Primers used for enrichment:
[0075] F: 5, -FAM-ATTCGACATGCCTGGACATA-3
[0076] R: 5, -biotin-TGCCTTGGTCATGTGGCATG-3
[0077] After measuring the OD260 of the oligomeric DNA library, centrifuge and dry; dissolve the library with 300ul binding buffer (containing 4.5g / L glucose in PBS, 5mMMgCl2, 2mg / mLBSA, 0.2mg / mL yeast tRNA), and take 250pmol (the first round of 10nmol ), 95°C for 5min, put it on ice quickly, and centrifuge quickly later.
[0078] Reverse screening: 250 pmol of the library was sucked into a culture dish with a diameter of 6 cm of human bone marrow mesenchymal stem cells with a growth coverage of 85%, supplemented to 1 mL with binding buffer; shaken at 4°C for 1.2 h; the supernatant was taken.
[0079] Positive sieve: Add the reverse sieve supernatant to a 6 cm culture dish with a g...
Embodiment 2
[0083] Example 2 Affinity Performance Verification
[0084] Take the target cell myeloma cell line XG7 and incubate with the aptamer GSL1-25 at different concentrations of 0, 25nM, 50nM, 100nM, 150nM, 200nM, 250nM, 300nM, 500nM, 800nM, 1000nM for 20min, wash and perform flow cytometry Experiments showed that as the concentration of the aptamer increased, its binding force to the target cells became stronger, and eventually the binding tended to be saturated. When the binding rate of the aptamer to the cell reaches 50%, the concentration of the aptamer required at this time is the equilibrium dissociation constant Kd, from which we can obtain the binding force constants of the aptamers to the cells as follows:
[0085]
[0086]
Embodiment 3
[0087] Adapter specificity analysis described in embodiment 3
[0088] 50nM FITC-labeled aptamer GSL1-25 was combined with target cells myeloma cell line XG7 or counter-screened bone marrow mesenchymal stem cells and oral epithelial cells for 20min, washed, fixed with 4% paraformaldehyde and confocal Microscopic experiments showed that for the myeloma cell line XG7, the aptamer GSL1-25 could well bind to it, and obvious fluorescence could be seen on the cell membrane, while for bone marrow mesenchymal stem cells and oral cavity For epithelial cells, none of the aptamers GSL1-GSL25 were obviously bound to it, so we could hardly see the fluorescence in the fluorescent field.
[0089] The following conclusions can be drawn through the above experiments: the aptamer sequences of the present invention can well bind to target cells, and the targeting is also very good.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com