Rna-guided gene editing and gene regulation
A gene and target gene technology, applied in the field of genome engineering and genome modification, genome engineering modification and genome modification of genes in muscles, and can solve the problems of inability to deliver macrodystrophin gene sequence, safety concerns, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0335] Materials and methods
[0336] Cell culture and transfection. HEK293T cells were obtained from the American Tissue Collection Center (ATCC) through the Duke University Cancer Center Facility (Duke University Cancer Center Facilities), in DMEM supplemented with 10% fetal bovine serum and 1% penicillin / streptomycin at 37 °C, 5% CO 2 down to maintain. According to the manufacturer's instructions, HEK293T cells were transfected with Lipofectamine2000 (Invitrogen). Transfection efficiencies were routinely greater than 80%, as determined by fluorescence microscopy following delivery of control eGFP expression plasmids. Transfect the Cas9 expression plasmid at a mass ratio of 3:1 to individual gRNA expression plasmids or an equal amount of gRNA expression plasmids (composed of equal amounts of a mixture of 4 gRNAs).
[0337] Primary mouse embryonic fibroblasts (PMEF-HL, Millipore, Billerica, MA) were seeded (75,000 / well) in 24-well TCPS plates (BD, FranklinLakes, NJ) in a...
Embodiment 2
[0352] result
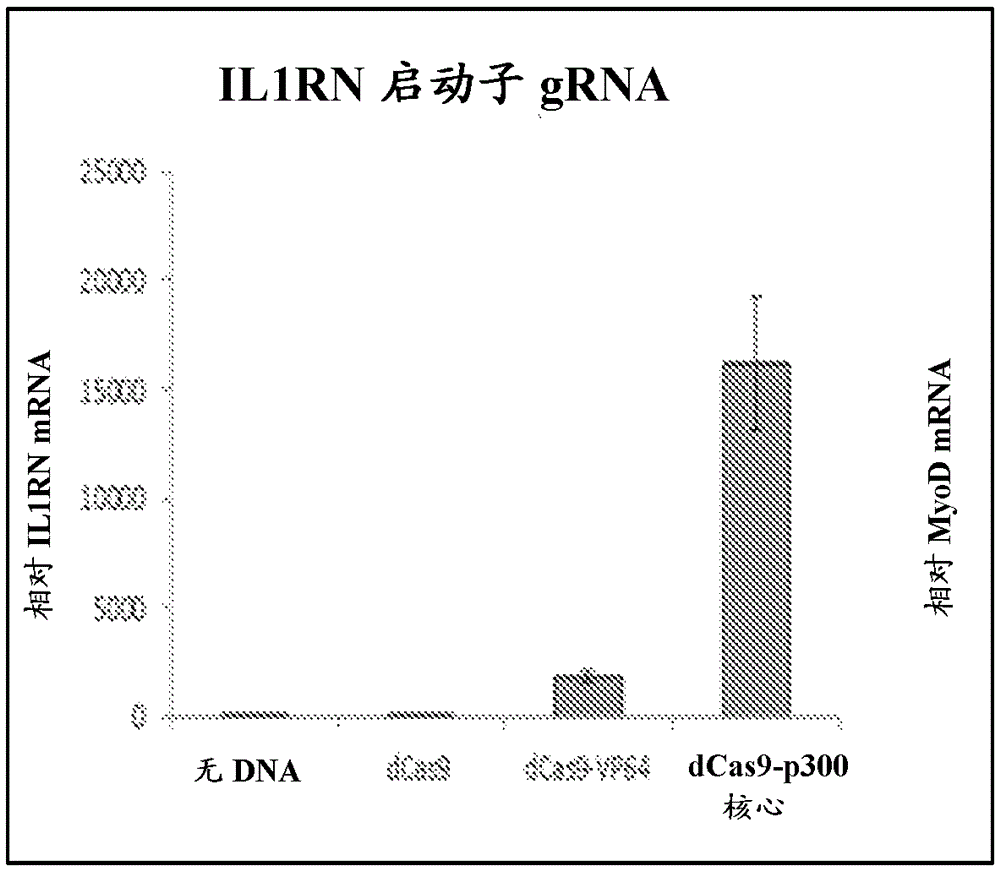
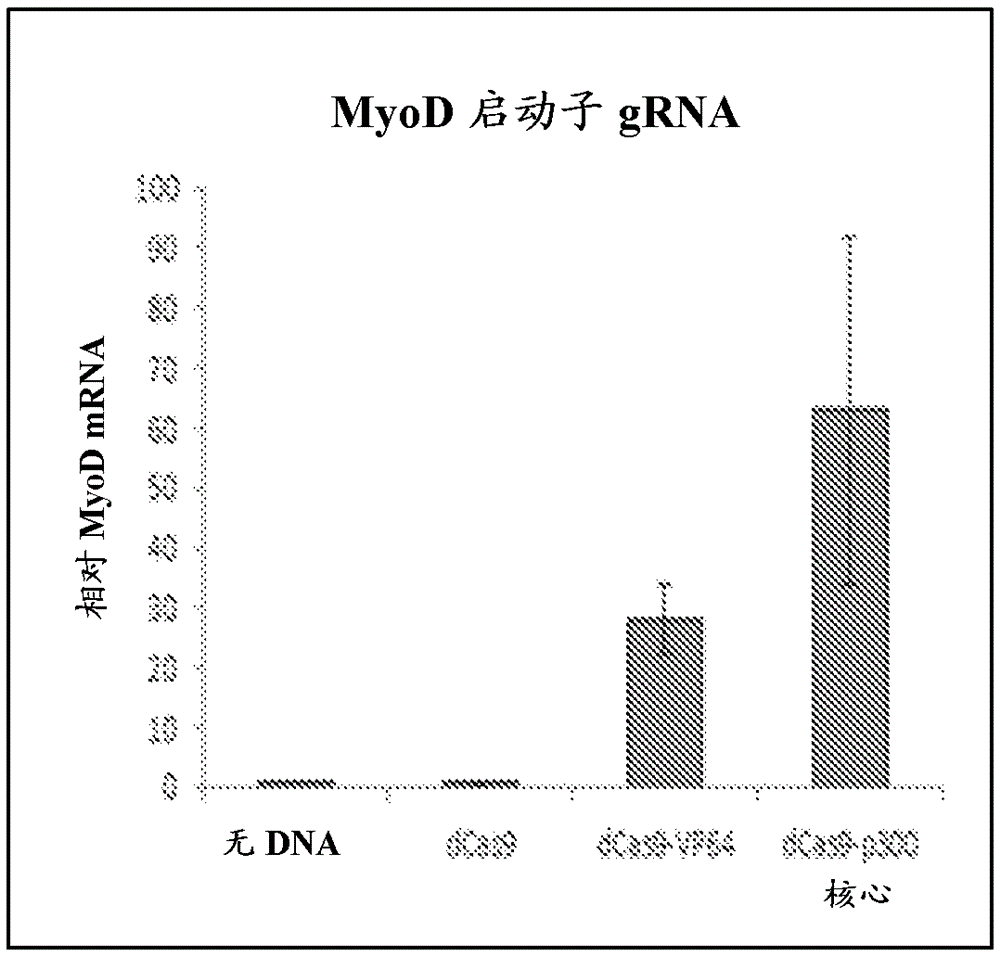
[0353] To generate a CRISPR / Cas9-based transcriptional activation system, the catalytic residues (D10A, H840A) of Cas9 were mutated to generate iCas9, which was genetically fused to the C-terminal VP64 acidic transactivation domain ( figure 1 a, b). Robust iCas9-VP64 expression was observed from transfected plasmids in human embryonic kidney (HEK) 293T cells by Western blotting of the N-terminal Flag epitope tag ( image 3 ). The CRISPR system recognizes its target by base pairing of a 20 bp sequence in the gRNA to a complementary DNA target followed by an NGG protospacer adjacent motif (PAM) sequence, where N is any base pair. Combination of synthetic transcription factors targeting endogenous human promoters results in synergistic and robust activation of gene expression. Therefore, within 500 bp of the transcription initiation site IL1RN Four gRNA target sites followed by the NGGPAM sequence were identified in the promoter of the gene ( Figure 4 ,Tab...
Embodiment 3
[0358] CRISPR targeting the dystrophin gene—methods and materials
[0359] plasmid construct . Expression cassettes of S. pyogenes sgRNA and human codon-optimized Cas9 (hCas9) nuclease were used as previously described (Perez-Pinera et al., NatMethods 10:973-976 (2013)). To generate a fluorescent reporter system to enrich CRISPR / Cas9-modified cells, GeneBlock (IDT) was synthesized. It contained a portion of the 3' end of the Cas9 coding sequence fused to the T2A skipping peptide immediately upstream of multiple cloning sites and was subsequently cloned into the hCas9 expression vector. The eGFP reporter gene was then cloned into a T2 vector for co-translation of Cas9 and eGFP proteins from the same expression vector (hCas9-T2A-GFP, SEQ ID NO: 116).
[0360] Cell Culture and Transfection . HEK293T cells were obtained from the American Tissue Collection (ATCC) via the Duke Cell Culture Facility and maintained in DMEM supplemented with 10% calf serum and 1% penicillin / stre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com