Construction method of penicillin-producing recombined strain of streptomyces virginiae IBL14
A technology for recombinant strains and construction methods, which is applied in the field of biomedicine and can solve problems such as no reports of penicillin recombinant strains
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
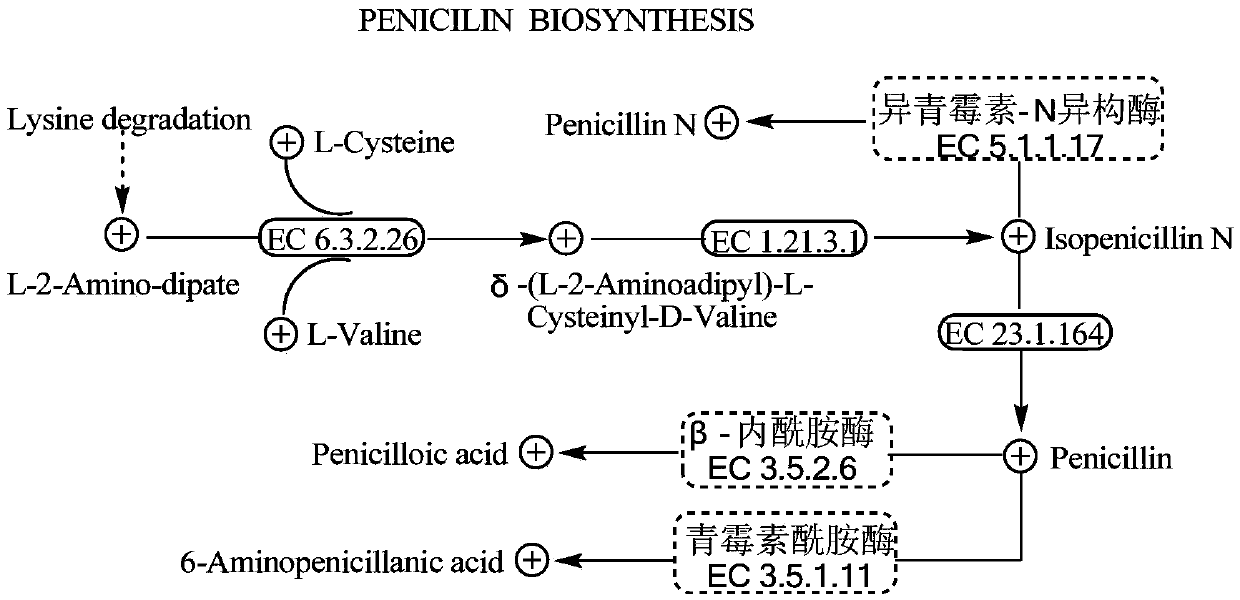
[0041] Example 1 (Isopenicillin-N isomerase gene sviIPI / GVGL005789 knockout in bacterial strain IBL-14)
[0042] (1) Primer design and DNA amplification of gene sviIPI01
[0043] According to the whole genome sequencing information, gene sviIPI-specific primers sviIPI-F and sviIPI-R were designed (Table 2). Genomic DNA of Streptomyces virginia IBL-14 was extracted, and PCR amplification of sviIPI gene was performed using PfuDNAPolymerase produced by Shanghai Sangon Bioengineering Co., Ltd., reaction conditions: 95°C for 5min, 94°C for 30s, 52°C for 30s, 72°C for 2min , 2.5U PfuDNAPolymerase (50μl reaction system) produced by Sangon, 30 cycles, 72°C for 10min. The PCR product was detected by 1% agarose electrophoresis, the kit was recovered, and the purified sviIPI full-length gene fragment was obtained for future use.
[0044] (2) Preparation of upstream and downstream homology arms
[0045] According to the complete sequence of the sviIPI gene (Table 1), the upstream homol...
Embodiment 2
[0059] Example 2 (Knockout of β-lactamase gene sviLT / GVGL000792 in bacterial strain IBL-14)
[0060] (1) Gene sviLT primer design and DNA amplification
[0061] The designed gene primers were sviLT-F and sviLT-R (Table 2). Using IBL-14 genomic DNA as a template, the sviLT gene fragment was amplified. Reaction conditions: 95°C for 5 min, 94°C for 30 s, 52°C for 30 s, 72°C for 3 min, 2.5 U of PfuDNA Polymerase produced by Sangon (50 μl reaction system), 30 cycles, 72°C for 10 min. The PCR product was detected by 1% agarose electrophoresis, the kit was recovered, and the purified sviLT full-length gene fragment was obtained for future use. Specifically with embodiment 1 step (1).
[0062] (2) Preparation of upstream and downstream homology arms
[0063] The amplification of the upstream and downstream homology arms is specifically the same as step (2) of Example 1. The designed upper and lower homology arm primers were sviLT-UF, sviLT-UR, sviLT-DF, sviLT-DR (Table 2). Using...
Embodiment 3
[0076] Example 3 (knockout of penicillin amidase gene sviPA / GVGL002963 in bacterial strain IBL-14)
[0077] (1) Gene sviPA primer design and DNA amplification
[0078] The designed gene primers were sviPA-F and sviPA-R (Table 2). Using IBL-14 genomic DNA as a template, the sviPA gene fragment was amplified. Reaction conditions: 95°C for 5 min, 94°C for 30 s, 55°C for 30 s, 72°C for 3.5 min, 2.5 U of PfuDNA Polymerase produced by Sangon (50 μl reaction system), 30 cycles, 72°C for 10 min. The PCR product was detected by 1% agarose electrophoresis, the kit was recovered, and the purified sviPA full-length gene fragment was obtained for future use. Specifically with embodiment 1 step (1).
[0079] (2) Preparation of upstream and downstream homology arms
[0080] The amplification of the upstream and downstream homology arms is specifically the same as step (2) of Example 1. The designed upper and lower homology arm primers are sviPA-UF, sviPA-UR, sviPA-DF, sviPA-DR (Table 2)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com