Aptamer with affinity to viral hepatitis C core antigen and application thereof
A nucleic acid aptamer, core antigen technology, applied in DNA/RNA fragments, measuring devices, instruments, etc., can solve the problems of high cost, harsh storage conditions, and high price of HCV virus, and achieve high application value, easy preservation, and easy storage. The effect of preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
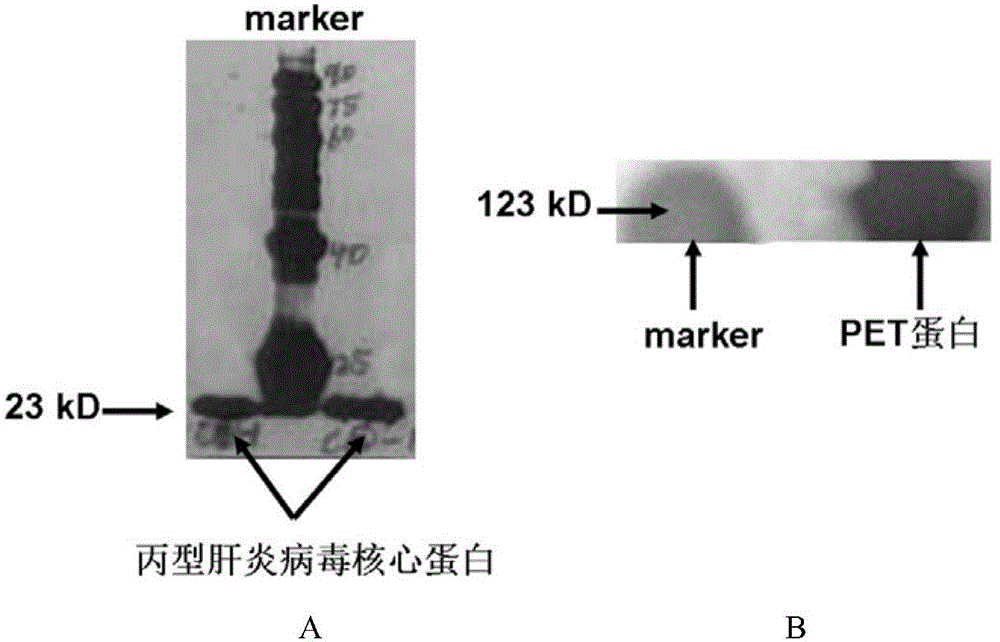
[0048] Embodiment 1, preparation of related proteins and related solutions
[0049] 1. Preparation of HCV core antigen (target protein) with histidine tag
[0050] 1. Amplification of the gene encoding HCV core antigen
[0051] Prepare the DNA shown in sequence 4 of the sequence listing (the coding gene of the hepatitis C virus core antigen, GENBANKACCESSIONNO.HM566118.1, the hepatitis C virus core antigen shown in the sequence 3 of the coding sequence listing), as a template for PCR amplification , using a primer pair consisting of primer 1 and primer 2 to perform PCR amplification to obtain a PCR amplification product.
[0052] Primer 1 (upstream primer): 5'-CGCGCGAATTCATGAGCACGAATCCT-3',
[0053] Primer 2 (downstream primer): 5'-CTGCAGGGATCCAGAGGCCGGGACGGTCA-3',
[0054] PCR amplification conditions: 95°C for 2min; 35 cycles of 95°C for 30s, 66°C for 30s, 72°C for 1min; 72°C for 7min.
[0055] 2. Construction of prokaryotic expression vector
[0056] ①Use restriction e...
Embodiment 2
[0085] Example 2, Screening and Preparation of Nucleic Aptamers
[0086] 1. Protein immobilization
[0087] 1. Take Ni-NTA agarose microbeads and place them in a 5ml centrifuge tube, remove the supernatant, and wash with PBS buffer three times;
[0088] 2. Disperse the microbeads in step 1 in the target protein (or control protein), incubate at room temperature for 1 hour, and centrifuge and wash with PBS buffer three times;
[0089] 3. Redisperse the microbeads from step 2 in 1ml of PBS buffer and store at 4°C for later use.
[0090] 2. Design of Random Nucleic Acid Library
[0091] A random nucleic acid library comprising 20 nucleotides at both ends and 40 nucleotides in the middle is designed as follows: 5'-ACGCTCGGATGCCACTACAG(N 40 ) CTCATGGACGTGCTGGTGAC-3'; N 40 Represents 40 random nucleotides, ie N is A, T, C or G.
[0092] 3. Screening of nucleic acid aptamers
[0093] 1. DNA library pretreatment
[0094] The random nucleic acid library is dissolved in binding buf...
Embodiment 3
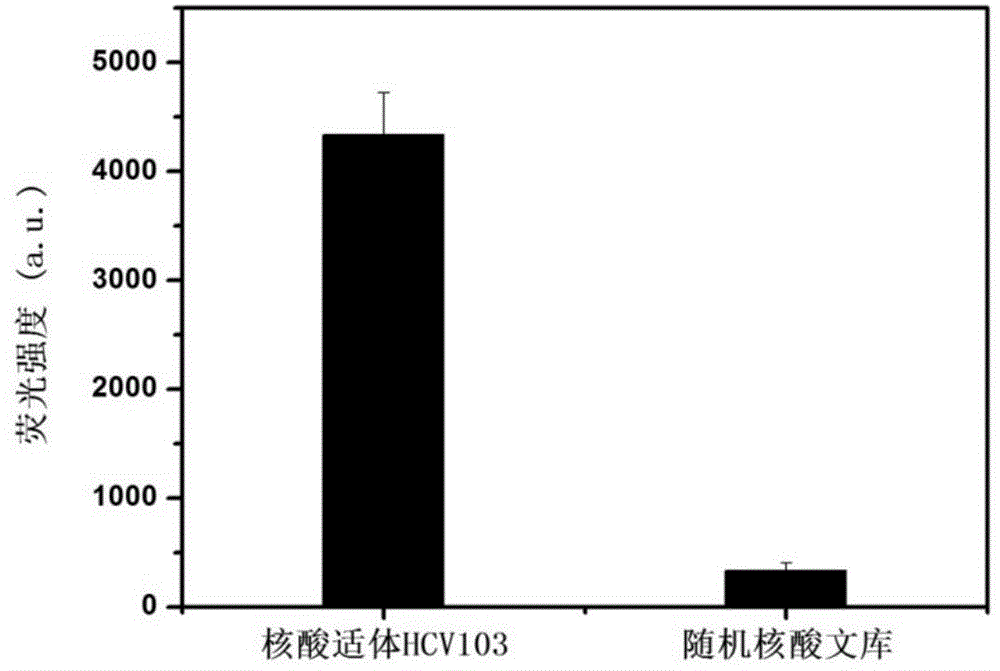
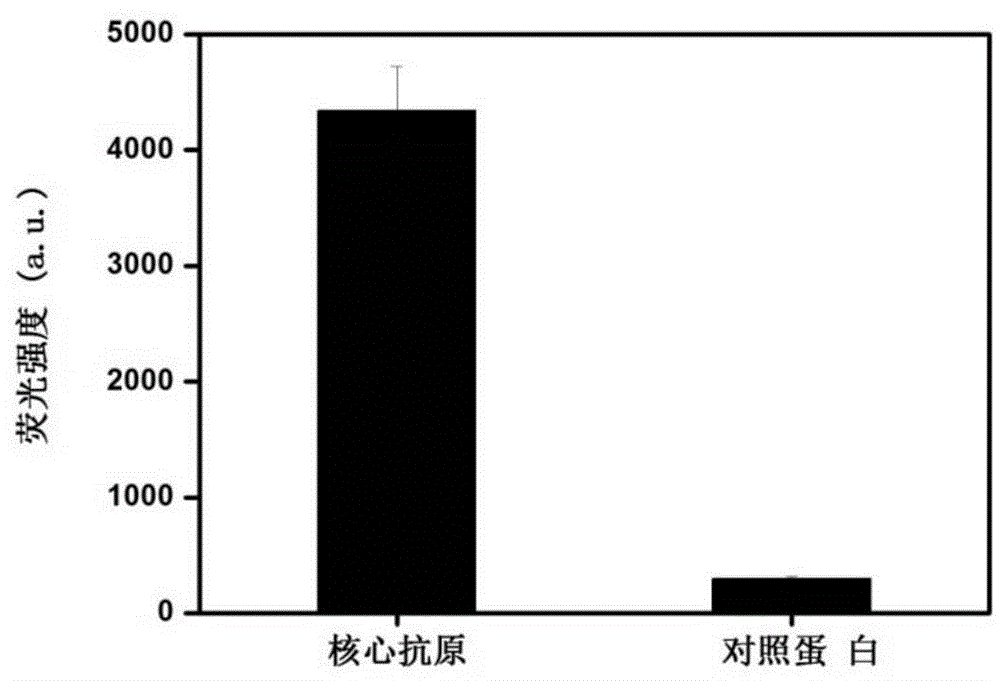
[0104] Example 3. Binding Characterization of Nucleic Aptamer and Hepatitis C Virus Core Antigen
[0105] 1. FITC labeling of nucleic acid aptamers and random nucleic acid libraries
[0106] The nucleic acid aptamer HCV103 prepared in Example 2 was labeled with fluorescein isothiocyanate (FITC).
[0107] The random nucleic acid library prepared in Example 2 was labeled with fluorescein isothiocyanate (FITC).
[0108] 2. Binding characterization of nucleic acid aptamer to HCV core antigen
[0109] 1. Immobilization of target protein
[0110] (1) Take 200 μl of Ni-NTA agarose beads and place them in a 5ml centrifuge tube, remove the supernatant, and wash with PBS buffer three times;
[0111] (2) Disperse the microbeads in step (1) in 1 mL of the target protein, incubate at room temperature for 1 h, and centrifuge and wash with PBS buffer three times;
[0112] (3) The microbeads in step (2) were redispersed in 1 ml of PBS buffer and kept at 4°C for use.
[0113] 2. Binding c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com