Primer pair, probe and kit for detecting bacterium MCR-1 gene
A technology of MCR-1 and primer pairs, which is applied in the field of probes and kits and primer pairs to detect bacterial MCR-1 drug resistance genes, can solve the problems of primers and probe kits that do not yet exist, and achieve accurate detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Embodiment 1 is used for detecting the kit of bacterial MCR-1 drug resistance gene and preparation method thereof
[0050] The kit for detecting bacterial MCR-1 drug resistance gene provided by the present invention consists of the following:
[0051] PCR reaction mix
[0052] The components and concentrations of the PCR reaction mixture are as follows: Buffer10×buffer 8 μL, upstream primer for detecting MCR-1 gene 0.6 μM, downstream primer for detecting MCR-1 gene 0.6 μM, MCR-1 probe 0.3 μM, amplification The upstream primer of the E.coli standard is 0.6 μM, the downstream primer of the amplified E.coli standard is 0.6 μM, the internal control probe is 0.3 μM, and each component of dNTP is 0.8 mM.
[0053] Wherein, in a specific embodiment, Buffer10× buffer is 10× ExTaqBuffer (Mg 2+ plus), 10×ExTaqBuffer (Mg 2+ plus) buffer was purchased from TAKARA Bao Biological Engineering (Dalian) Co., Ltd., model 9152A.
[0054] The upstream primer sequence for detecting the M...
Embodiment 2
[0063] Example 2 is used to detect the use method of the kit of bacterial MCR-1 drug resistance gene
[0064] 1. Sample DNA extraction:
[0065] The kit used is the OMEGA Bacterial Genome Extraction Kit. For specific steps, please refer to the instruction manual. The contents of the instruction manual are as follows:
[0066] (1) Take 1 mL of bacterial culture solution or bacteria, and centrifuge at 10,000×g for 1 min.
[0067] (2) Remove the supernatant, add 250 μL solution I (containing RNaseA), and vortex until the cells are completely suspended.
[0068] (3) Add 250 μL of solution II, and gently invert the centrifuge tube 4-6 times to obtain a clear lysate. It is best to incubate at room temperature for 2 minutes. Vigorous mixing will shear the chromosomal DNA and reduce the purity of the plasmid. (Store solution II should be tightly capped).
[0069] (4) Add 350 μL of solution III, mix gently by inverting several times until white flocculent precipitates appear, and c...
Embodiment 3
[0089] Example 3. Sensitivity analysis for detecting bacterial MCR-1 drug resistance gene kit
[0090] 1. Preparation of reference DNA nucleic acid
[0091] Construction of a plasmid containing the MCR-1 gene
[0092] The MCR-1 gene (SEQ ID NO.7) as described in the summary of the invention was synthesized by GenScript. The synthetic gene was connected to the pUC57 vector (provided by GenScript), a plasmid containing pUC57-MCR1 was constructed, and sequenced for proofreading.
[0093] 2. Sensitivity and specificity analysis of the kit for detecting MCR-1 drug resistance gene
[0094] Preparation of reference substances at different concentrations
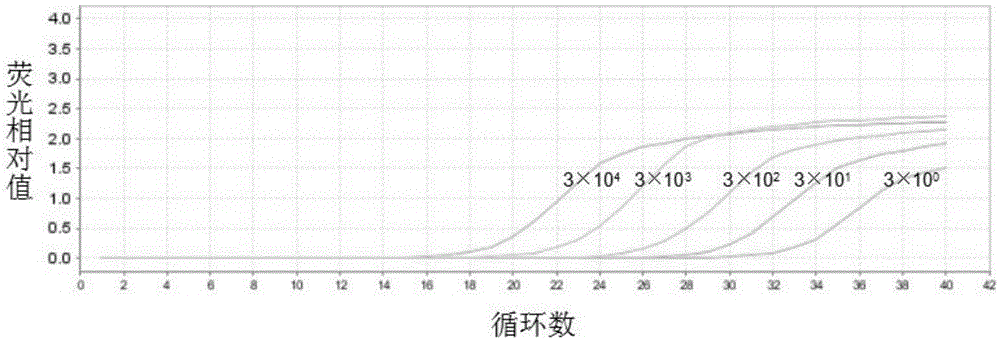
[0095] Quantify the plasmid containing the MCR-1 gene synthesized in the above steps, and quantify it as 3×10 10 copies / μL, and serially diluted to obtain 3×10 5 copies / μL, 3×10 4 copies / μL, 3×10 3 copies / μL, 3×10 2 copies / μL, 3×10 1 copies / μL, 3×10 0 Copy / μL and other different concentration gradient dilutions.
[0096] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com