Traceable non-fluorescence-marked escherichia coli and preparation method thereof
A technology of Escherichia coli and recombinant Escherichia coli, which is applied in the field of bioengineering, can solve problems such as uncontrollable insertion sites, achieve the effect of convenient tracking and traceability, and reduce impact
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
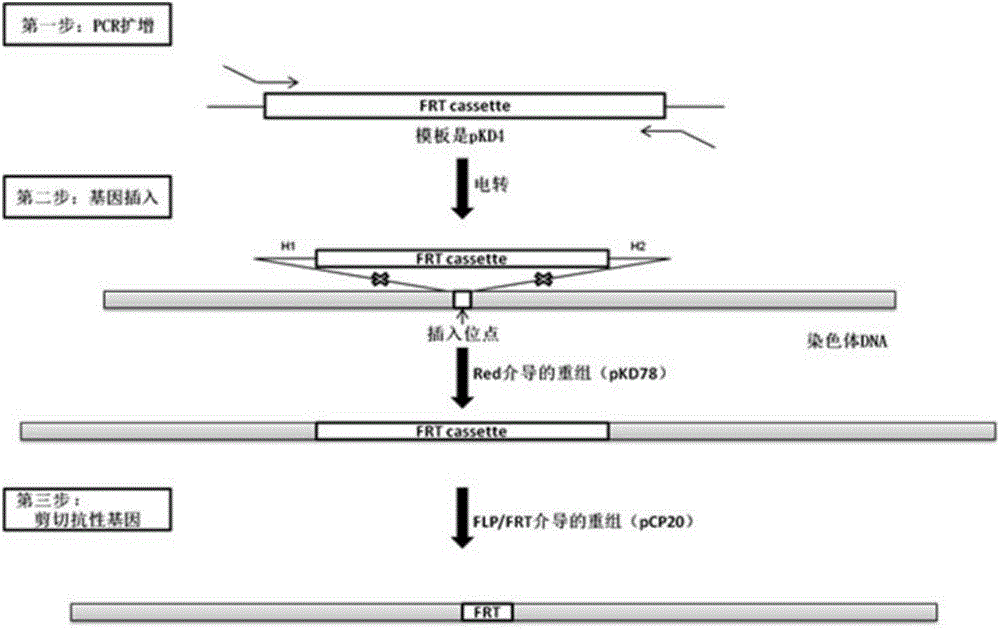
[0020] The embodiment of the present invention provides a method for preparing traceable non-fluorescence-labeled Escherichia coli. For the schematic diagram of the method, see figure 1 , including the following steps:
[0021] S10, using pKD4 as a template, the FRTcassette product containing a sequence homologous to the manX gene is amplified by PCR with pKD4 amplification primers;
[0022] S20, transforming the pKD78 plasmid into Escherichia coli to be transformed, and obtaining Escherichia coli containing the pKD78 plasmid;
[0023] S30, transforming the FRTcassette product into Escherichia coli containing the pKD78 plasmid and performing site-directed recombination with the chromosome of Escherichia coli to form recombinant Escherichia coli;
[0024] S40, the kanamycin resistance gene in the recombinant E. coli chromosome FRTcassette is removed by the suicide plasmid pCP20, and the traceable non-fluorescence-labeled E. coli of the present invention is obtained.
[0025] ...
Embodiment 1
[0041] S10, using pKD4 as a template, using the above-mentioned upstream primer HP1 and downstream primer HP2 to amplify to obtain a FRTcassette product with a size of about 1600 bp containing a sequence homologous to the Escherichia coli manX gene.
[0042] Specific PCR reaction system (50μl):
[0043]
[0044] PCR process:
[0045]
[0046]
[0047] After the PCR process, the FRTcassette product was purified and recovered by 1% agarose gel electrophoresis for future use.
[0048] S20, transforming the pKD78 plasmid into Escherichia coli to be transformed to obtain Escherichia coli containing the pKD78 plasmid, the detailed implementation steps include:
[0049] S21, preparing competent E. coli, the specific method steps can be carried out with reference to the steps introduced in the "Molecular Cloning Experiment Guide" (third edition); however, the cell suspension in the one-step method adopts the following components specifically developed by the present inventio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com