Molecular biological method for quickly identifying Budorcas taxicolor
A technology of molecular biology and species identification, which is applied in the field of molecular biology for rapid identification of takins, can solve the problems of distinguishing and identifying takins, and achieve the effect of saving time and reducing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
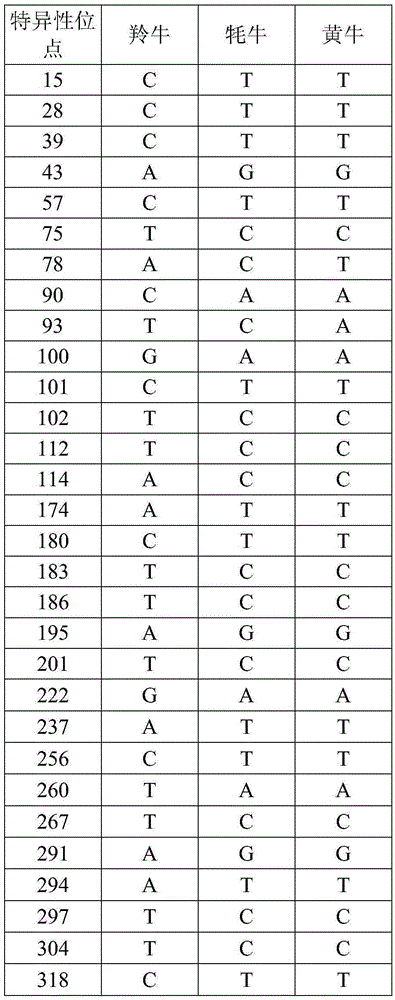
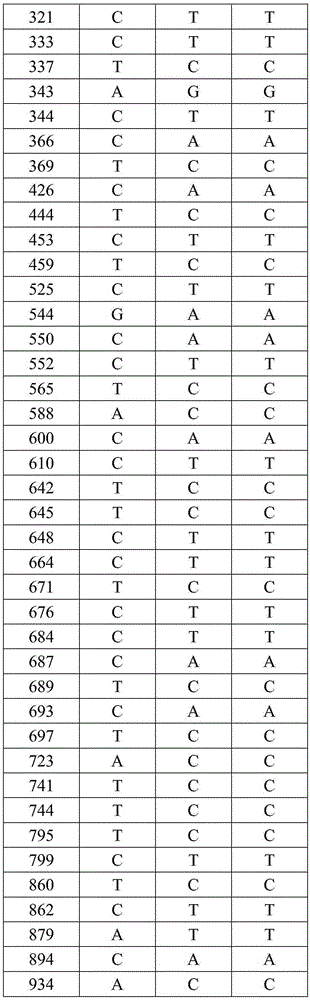
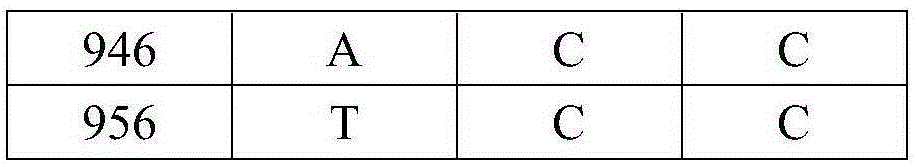
[0024] 1. Use phenol-chloroform method or DNA extraction kit to extract the total DNA of takin, yak and cattle. In order to avoid contamination between animal residues and humans, the superficial muscle was removed, and DNA was extracted from fresh, uncontaminated muscle. DNA extraction refers to the standard phenol-chloroform method (thestandardphenol / chloroform protocol). Using DNA barcode primers to PCR amplify the mitochondrial Cytb gene fragments and sequence determination of takin, yak, and cattle, specifically comprising the following steps:
[0025] 1) The PCR reaction system is 45 μL, including: 1 μL of DNA template with a concentration of 50 ng, Mix 20 μL, 1.0 μL of upstream and downstream primers with a concentration of 10 pM, and add water to make up 45 μL of the system, in order to prevent the evaporation of the liquid in the PCR tube during the reaction , add 2.0 μL glycerol to the surface of the system; the reaction is completed on a MyCyclerThermalCycler therm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com