Method for directly detecting miRNA in absolute quantification mode
An absolute quantitative and direct technology, applied in the field of plasma miRNA quantitative detection, can solve the problem of inability to measure miRNA, and achieve the effects of improving sensitivity and repeatability, reducing method errors, and reducing the range of measurement variation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1: Effects of different concentrations of SDS, TritonX-100 and Tween-80 etc. on miRFLP assay
[0049] Many surfactants, such as SDS, Tween-20, TritonX-100, etc., can promote DNA synthesis at low concentrations, and are often used to increase the efficiency of RT or PCR reactions. High concentrations of surfactants inhibit the efficiency of enzyme reactions, but can also help dissolve cell membrane components and dissociate the tertiary structure of proteins. The inventors digested H1299 cells reaching 90% saturation in a 100 mm culture dish with trypsin, washed with PBS and divided them into 6 equal parts, added PBS, 0.5% SDS, 1% TritonX-100, 0.5% Tween-80, and carried out crack. After heating at 75°C for 5 minutes, quickly cool on ice, mix well and centrifuge at 10,000 g for 10 minutes, take the supernatant, and use miRFLP assay to measure miR-15a / b, miR-155, and miR-222. Dilute the sample so that the concentration of surfactant contained in each RT reaction ...
Embodiment 2
[0050] Example 2: Effect of 3' end-modified adapter oligonucleotide chains on miRFLP assays
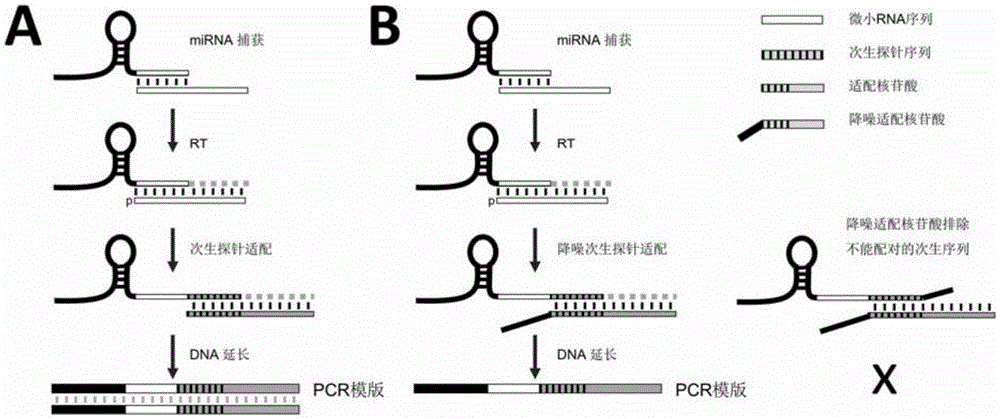
[0051] Using the miRFLP assay to detect miR-150 / 146a / 222 / 146b, the adapter oligonucleotide chain used for cDNA elongation was used to add "AATTTAA" to the 3' end of the adapter oligonucleotide chain for three The results are listed in Table 1. Fluorescence intensity readings obtained with adapter oligonucleotides with modified 3' ends are generally lower than those obtained with adapter oligonucleotides with unmodified 3' ends. The modified adapter oligonucleotide chain has no primer function, which reduces the number of non-specific PCR templates and better maintains the linear relationship determined by the method. The detection of fluorescence intensity can be improved by increasing the number of PCR cycles or reducing the dilution factor to make up for the decrease in the detection limit caused by the decrease in the PCR template. Such as figure 1 As shown in B, mature miRNAs c...
Embodiment 3
[0054] Embodiment 3: The performance analysis of extracting-free miRFLP direct assay
[0055] The operation process of the comprehensively improved extraction-free miRFLP direct assay is as follows: the target miRNA to be tested is mixed evenly with the dynamic miRNA standard, after miRNA reverse transcription, cDNA tailing modification, biotinylation with agarose streptomycin coupling reagent The labeled omega products are enriched and purified, and then amplified simultaneously by fluorescent PCR. Finally, DNA sequencer is used to analyze the length of DNA fragments and quantitative fluorescence:
[0056] RNA denaturation solution: 5 μl 1% Tween-80, 200 μl 5xRT buffer, 100 μl ddH 2 O; dynamic molecular standard mixture; 2 μl RNA dynamic molecular standard (including: standard1RNA (dynamic molecular standard 1, and so on): 120,000; standard2RNA: 20,000; standard3RNA: 3340; standard4RNA: 600, the RNA sequence of the dynamic molecular standard See Table 12, its base sequences ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com