A sgRNA base mismatch target site library and its application
A target site and library technology, applied in the field of gene modification, can solve the problems of fixed sequence recruiting nuclease efficiency changes, recruiting nuclease efficiency changes, difficult to evaluate recognition efficiency and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
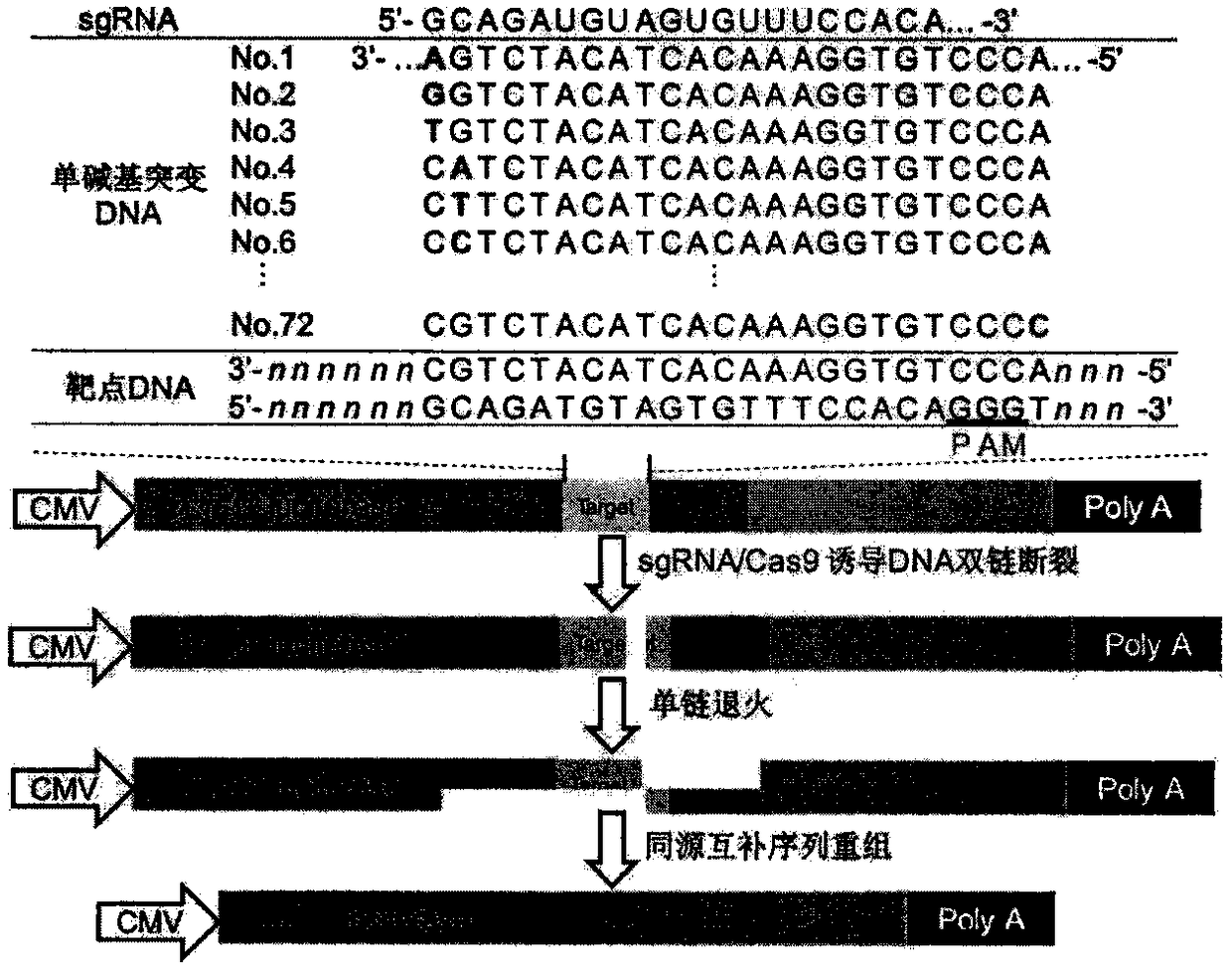
[0035] Example 1. sgRNA off-target detection system and sgRNA single-base mismatch target site library construction
[0036] The sgRNA off-target detection system used in the present invention is the pSG-target Cloning Kit & Single Stranded Annealing Assay (BV3100) system provided by Biomics Biotechnology Co., Ltd. (http: / / www.biomics.com). The system consists of three plasmids: the pCas plasmid encodes the cas9 protein, the psgRNA plasmid encodes the sgRNA sequence, and the pTarget plasmid encodes an inactive firefly luciferase reporter gene. In the inactive state, the firefly luciferase reporter gene is divided into two parts separated by a stop codon, a target site for sgRNA, and two 1000-base direct repeat sequences. When the sgRNA / Cas9 complex acts on the target site of the sgRNA to produce a double-strand cut, the DNA repair system in the cell will, on the basis of the direct repeat sequences on both sides of the cut site, through homologous recombination, convert the br...
Embodiment 2
[0098] Embodiment 2, sgRNA activity detection
[0099] Detection of sgRNA activity: The day before the experiment, HEK293 (human embryonic kidney cells) was mixed with 2.5×10 4 The density of cells / well was inoculated into a 48-well cell culture plate, and the medium was DMEM medium containing 2mM L-glutamine, 10% fetal bovine serum (Sigma), 100U / ml penicillin and 100ug / ml streptomycin. When the cells grew to 30-50% density, the cell transfection experiment was performed using Lipo2000 (invitrogen) transfection reagent. According to the requirements of the transfection reagent instructions, each culture well was transfected with 200ng of pCas plasmid, 200ng of psgRNA plasmid, 30ng of pTarget plasmid, and 5ng of renilla luciferase control plasmid pRL-TK. Incubate the cells at 37°C CO 2 After culturing in the incubator for 24 hours, the dual luciferase reporter gene detection system (Promega, Reporter Assay System) and a microplate reader (Synergy HT, BioTek, USA) were used ...
Embodiment 3
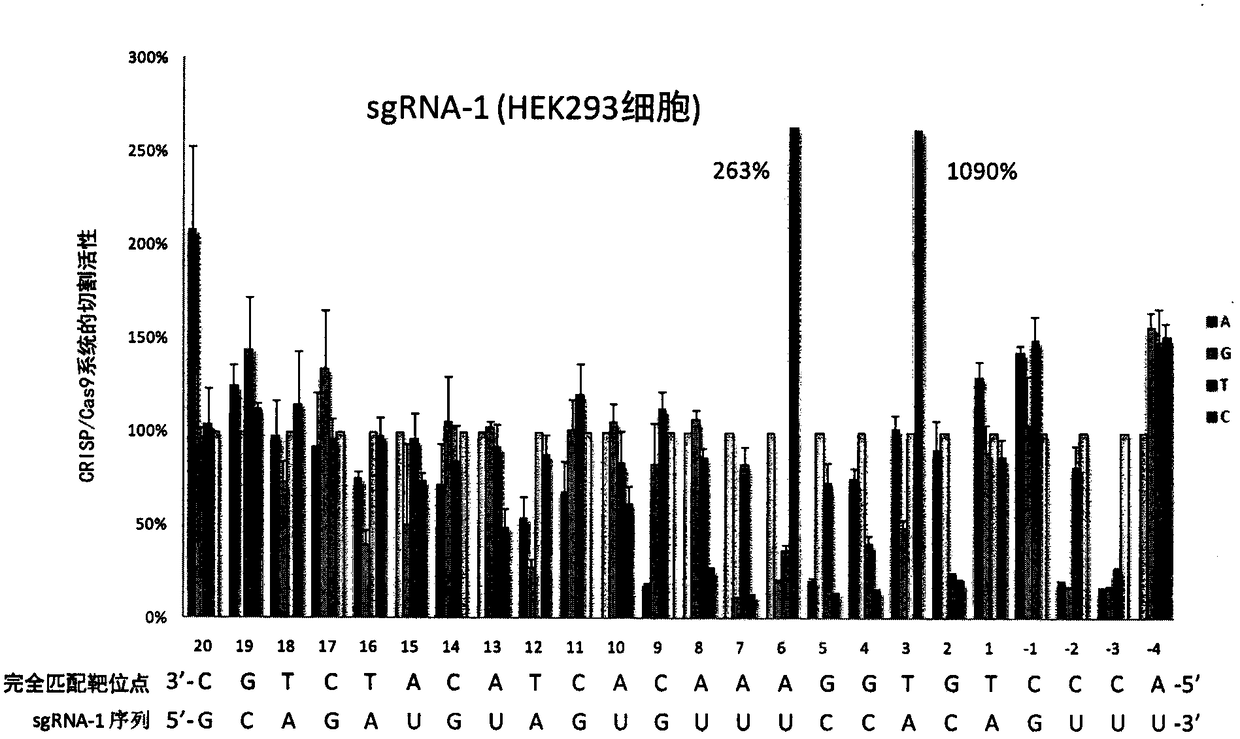
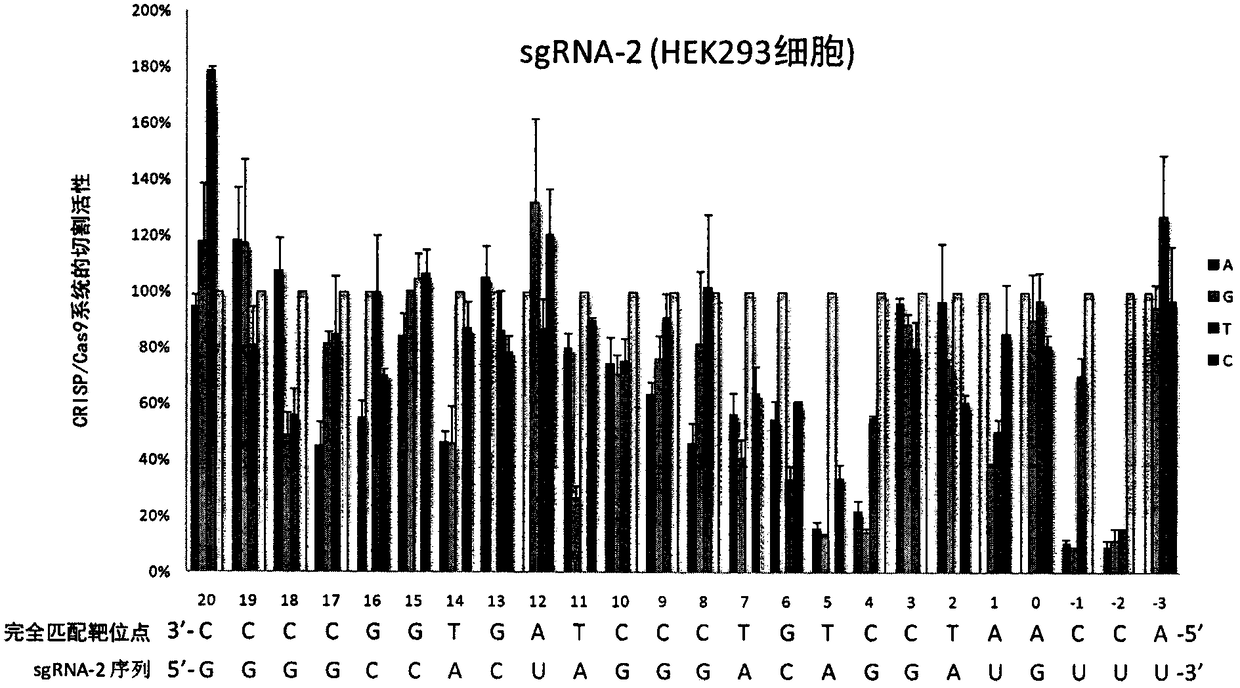
[0100] Example 3. Application of single-base mismatch target site library in the study of sgRNA off-target effects
[0101] Using the method described in Example 2, in cultured HEK293 cells, using the single-base mismatch target site sequence library constructed in Example 1, the off-target effects of two sgRNAs were detected. The experimental results are as follows figure 2 , 3 shown.
[0102] The above experimental results show that using the single base mismatch target site library provided by the present invention can systematically detect the off-target effect when the sgRNA and the target site sequence are mismatched at each position.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com