A strain of Streptomyces clavulatum with high activity clavulanate aminoacetylase and its application
A technology of acid aminoacetylase and Streptomyces clavulatus, which is applied in the field of microbial breeding and biology, can solve the problem of difficult selection of genetic modification sites, and achieve the effects of reducing randomness and blindness, high efficiency, and strong pertinence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Obtaining target genes containing mutated bases by continuous error-prone PCR
[0034] Mutation of the clavulanate aminoacetylase gene in vitro using sequential error-prone PCR. Specifically:
[0035] 1. The primers for error-prone PCR are:
[0036] Sequence of upstream primer GcasForward: GCTCTAGAGCCCGCAGTGTGATGAAG;
[0037] Downstream primer GcasReverse sequence: GCGAATTCGGGTGGCGTCTCCGCTCACG.
[0038] 2. PCR system: template DNA 2ul, PCR buffer 5ul, Taq DNA polymerase 1ul, dATP 1~3ul, dTTP1~3ul, dGTP 1~3ul, dCTP 1~3ul, upstream primer 2ul, downstream primer 2ul, 25mM MgCl 2 1~8ul, 5mMMnCl 2 0~1ul, add ultrapure water to 50ul.
[0039] 3. PCR conditions: pre-denaturation at 97°C for 10 minutes, denaturation at 97°C for 30 seconds, annealing at 52-62°C for 30 seconds, extension at 72°C for 2 minutes, 35 cycles, and final extension for 10 minutes.
[0040]The template DNA for the first PCR was derived from the industrial strain Streptomyces clavulus B02,...
Embodiment 2
[0042] Example 2: Construction of a library of clavulanic acid aminoacetylase mutants from Streptomyces clavulanate
[0043] The continuous error-prone PCR products in Example 1 were double-digested with XbaI and BamHI, and the digested products were recovered after gel agarose electrophoresis separation, and combined with the same double-digested shuttle vector pSET152 (pSET152 is a prior art Conventional plasmids) were ligated, transformed into Escherichia coli DH5a, and the transformants were picked to extract the plasmids for double enzyme digestion identification.
[0044] Using conjugative transfer technology, the recombinant pSET152 was transferred into the clavulaminic acid aminoacetylase gene deletion strain of Streptomyces clavulinus strain B02 (the gene deletion strain was obtained by conventional gene knockout technology based on strain B02), and a series of Zygotes to construct a mutant library of the clavulanate aminoacetylase gene.
Embodiment 3
[0045] Example 3: Plate screening of zygotes
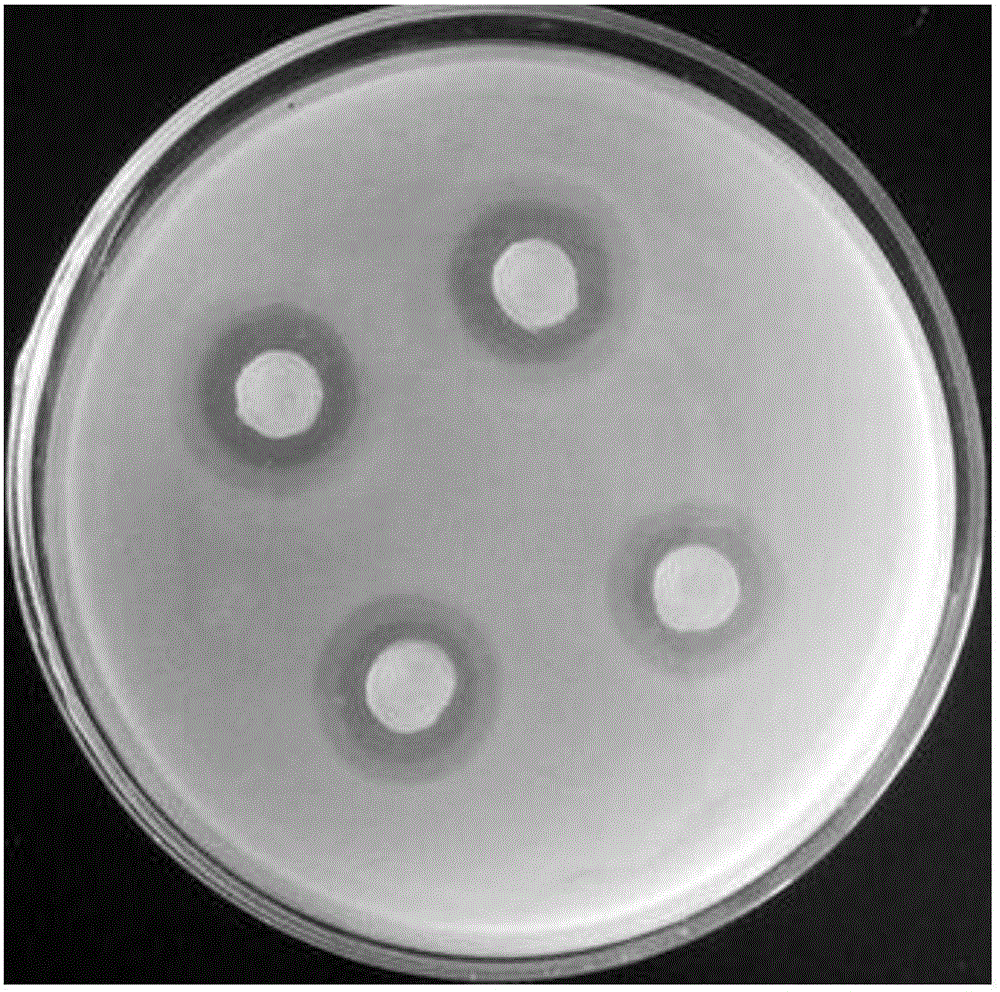
[0046] Escherichia coli with β-lactamase was used as an indicator bacterium, and ampicillin-containing LA was used to make a screening plate, and the Streptomyces clavulatus zygote (prepared in Example 2) was screened by the plate transparent circle method. First, 6897 zygotes were transferred to BSCA solid plates and cultured at 25°C for 6 days, then excavated zygote agar blocks with a diameter of 6 mm, placed on a screening plate, cultured at 37°C for 16 hours, and measured the diameter of the transparent circle (see figure 1 ). At the same time, strain B02 was used as a control. The larger the diameter of the transparent circle, the greater the ability to synthesize clavulanic acid. A total of 564 positive mutant strains were selected, which were continued to be cultured until sporulation and were numbered B02-1 to B02-564.
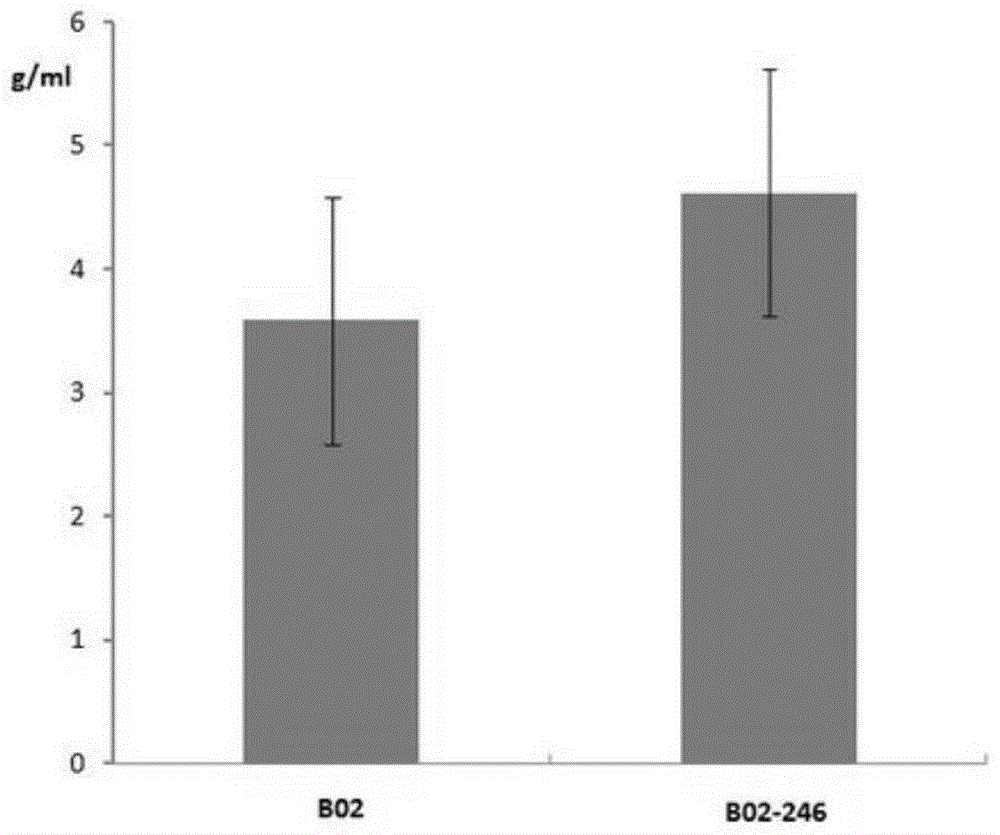
[0047] The spores of 564 positive mutants were diluted to 10 5 1 / ml, take 100ul and spread on BSCA so...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com