Creatinase mutant with increased thermal stability
A technology of creatine hydrolase and thermal stability, which is applied in the direction of hydrolytic enzymes, enzymes, and microbial-based methods, and can solve problems such as unsatisfactory thermal stability and decreased enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] The acquisition of embodiment 1 high thermostability mutant strain
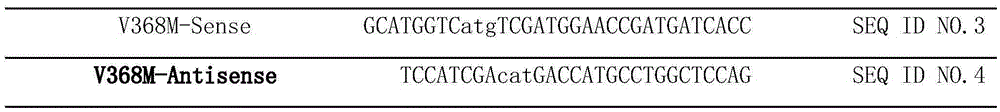
[0023] Using the site-directed mutagenesis kit (TaKaRa), design a pair of primers (as shown in Table 1), and use the constructed pET20J as a template to perform PCR to mutate the 358th valine inside the creatine hydrolase molecule to methionine , named V368M, the PCR reaction conditions were 98°C for 3min, 34 cycles (98°C for 3min, 58°C for 30S, 72°C for 1min30S), and 72°C for 10min. PCR amplification system: template 1 μl, upstream and downstream primers 1 μl, 2xPrimeStar 24 μl, sterilized double distilled water 24 μl. After PCR, add 5 μl of FDbuffer and 1 μl of DpnI to digest for 1 hour. The digested PCR product was purified, recovered and transformed using a gel recovery kit. Transformants were sequenced by Shanghai Sangon. The correctly sequenced transformant was named Pet20J-V368M.
[0024] Table 1
[0025]
Embodiment 2
[0026] Example 2 Verification of High Thermostable Creatine Hydrolase Mutant Strain
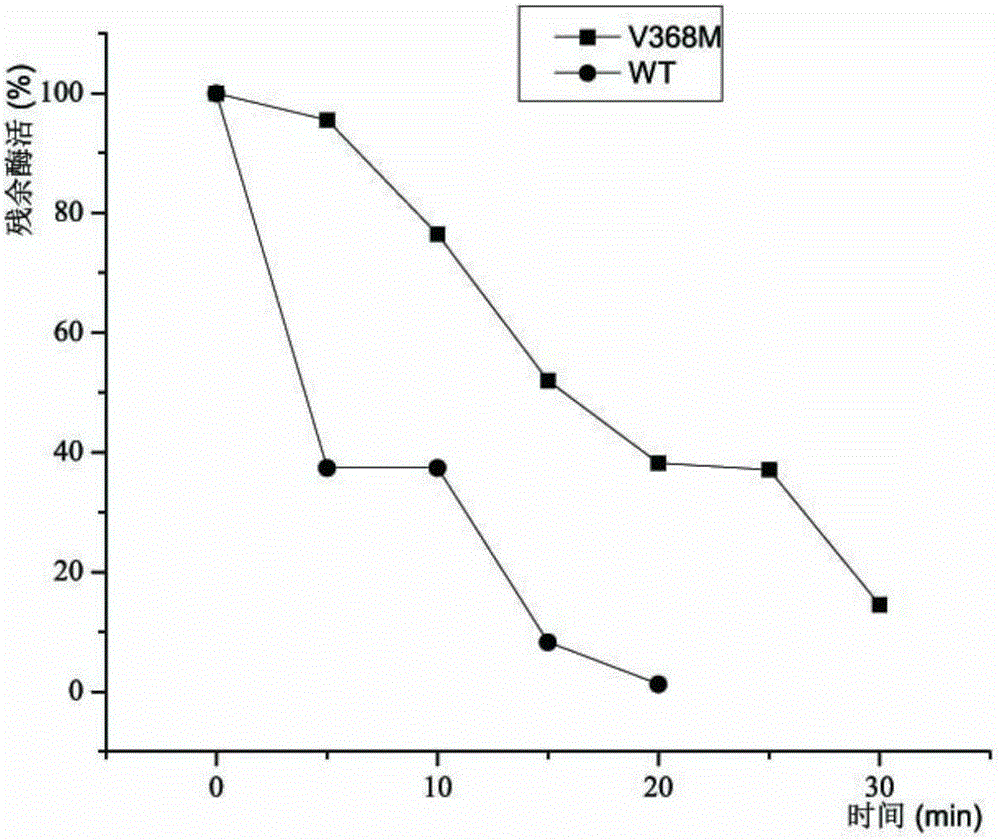
[0027] The plasmid with correct sequencing was transformed into E.coliBL21, and the transformants were selected and inoculated into LB liquid medium, cultured at 37°C for 12 hours, and transformed into TB medium with an inoculum size of 3%. When the bacteria grew to OD600 of 3, IPTG was added to make the final concentration 0.6mmol / L, and the culture temperature was lowered to 30°C for 8h. Collect the fermented bacteria. After ultrasonic crushing, the enzyme activity was measured at the same temperature for different time.
Embodiment 3
[0028] The purification of embodiment 3 creatine hydrolase
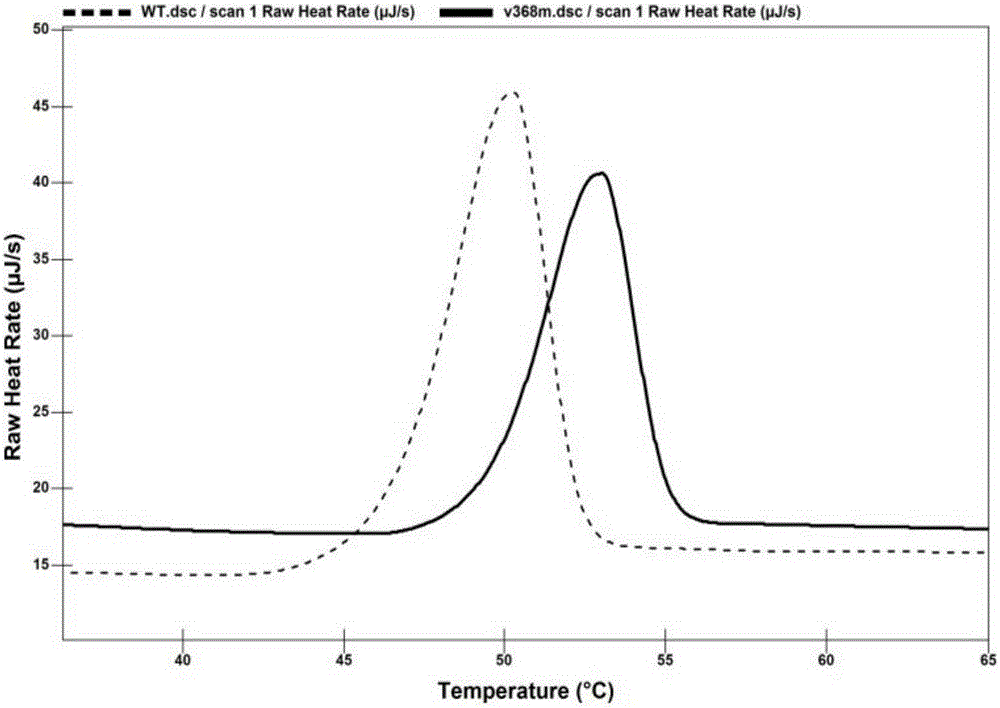
[0029] After crushing Escherichia coli, carry out ammonium sulfate precipitation, select the precipitation with 55%--75% ammonium sulfate saturation, dissolve the protein precipitate with a small amount of phosphate buffer (pH7.0), and dialyze for 24 hours to remove ammonium sulfate in the enzyme solution. According to the isoelectric point properties of creatine hydrolase, the QFF column was selected for ion exchange purification. The QFF column was equilibrated with phosphate buffer for 30 min, the crude enzyme solution was injected into the purification column, and the target protein was eluted with phosphate buffer containing 1M NaCl.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com