Cattle muscle enhanser and application thereof
A technology of bovine muscle and subsequence, applied in bovine muscle enhancer and its application field, can solve the problem of reducing the safety of transgenic cattle and achieve the effect of enhancing transcriptional activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1 PCR amplification bovine muscle enhancer sequence
[0036] Genomic DNA of Angus cattle was extracted and used as a template (concentration: 80ng / ul); PCR amplification was performed with Q5 ultra-high-fidelity DNA polymerase (NEB, M0491L) and specific amplification primers to obtain beef muscle enhancement Subsequence (its nucleotide sequence is shown in SEQ ID No.1).
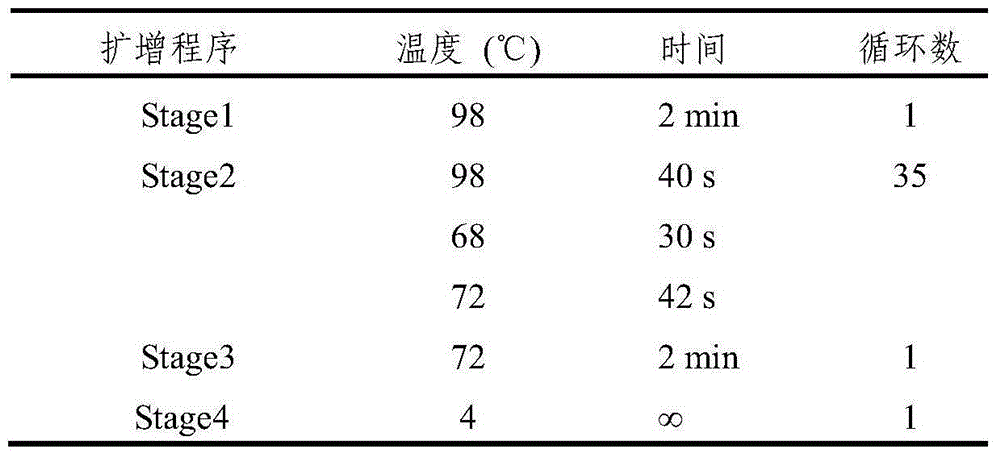
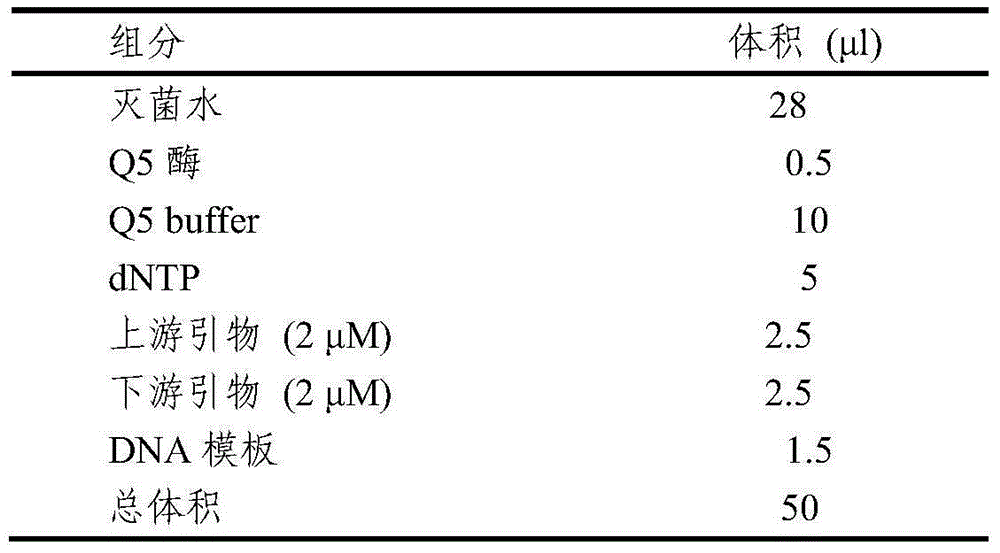
[0037] The PCR amplification program is shown in Table 1, and the PCR reaction system is shown in Table 2.
[0038] The specific amplification primer pair is as follows:
[0039] Upstream primer sequence (containing BamHI restriction site and protective base): ATCGGGATCCGCCAAGATAAGGTTGGGACA;
[0040] Downstream primer sequence (containing SalI restriction site and protection base): ATCGGTCGACTTTAAAACCCCCTCACCCTTC.
[0041] Table 1 PCR amplification program
[0042]
[0043] Table 2 PCR reaction system
[0044]
Embodiment 2
[0045] Example 2 Construction of the PGL3 luciferase reporter carrier containing the enhancer sequence of the present invention
[0046] 1) Digest the enhancer sequence amplified in Example 1 and the pGL3-promoter vector (PromegaE1761) with BamHI and SalI restriction endonucleases;
[0047] 2) After electrophoresis, cut the gel to recover the digested product;
[0048] 3) The enhancer sequence and the vector were ligated overnight at 4°C with T4 ligase (NEBM0202L);
[0049] 4) Use the ligated product to transform Escherichia coli, and after plating, incubate upside down at 37°C for about 12 hours;
[0050] 5) Pick a single clone, streak it on an LB culture plate, incubate it upside down at 37°C for 8 hours, and carry out bacterial cell PCR identification. Detection of amplification results by PCR and agarose electrophoresis;
[0051] 6) Perform DNA sequencing on PCR-positive colonies;
[0052] 7) Transfer the expected colonies to LB medium and culture them on a shaker at 3...
Embodiment 3
[0054] Example 3 Functional Verification of Bovine Muscle Enhancer
[0055] 1. When the confluence of C2C12 cells is 60%-70%, use liposome 2000 (11668-019, Invitrogen) to co-transform pGL3-1357 and pRL-TK (E2241), pGL3-promoter and For pRL-TK, when the confluence of the cells reaches 90%, the cells are cultured with a differentiation medium, and the cells are induced to differentiate for 6 days.
[0056] The differentiation medium is DMEM high glucose medium containing 2% horse serum and 1% penicillin / streptomycin.
[0057] The conditions for culturing with differentiation medium were: 37°C, 5% CO2.
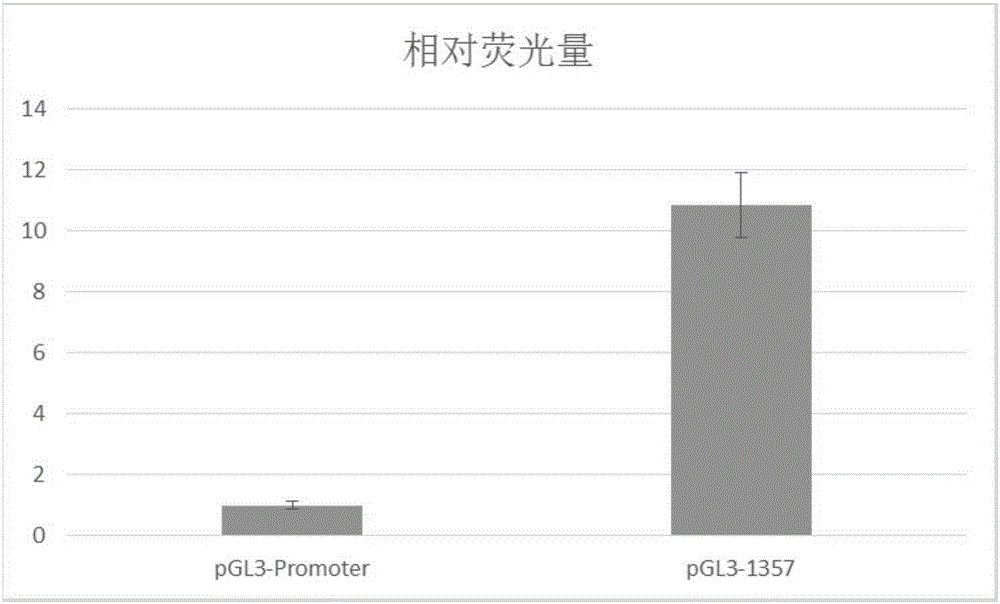
[0058] 2. Use after 6 days of differentiation The Luciferase assay (PromegaE2920) kit detects firefly luciferase and sea cucumber luciferase, and uses the ratio of firefly luciferase / sea cucumber luciferase to illustrate whether the enhancer works, such as figure 1 As shown, the enhancer discovered in the present invention has about 11-fold enhancement effect.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com