Microbial growth phenotype predication method based on control-metabolic network integration model
A metabolic network and model prediction technology, applied in the field of joint modeling of gene regulation network and metabolic network, can solve the problem of less research on the influence of metabolic phenotype, and achieve the effect of improving accuracy and good growth phenotype
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0019] The technical solutions of the present invention will be further described below in conjunction with the accompanying drawings and embodiments.
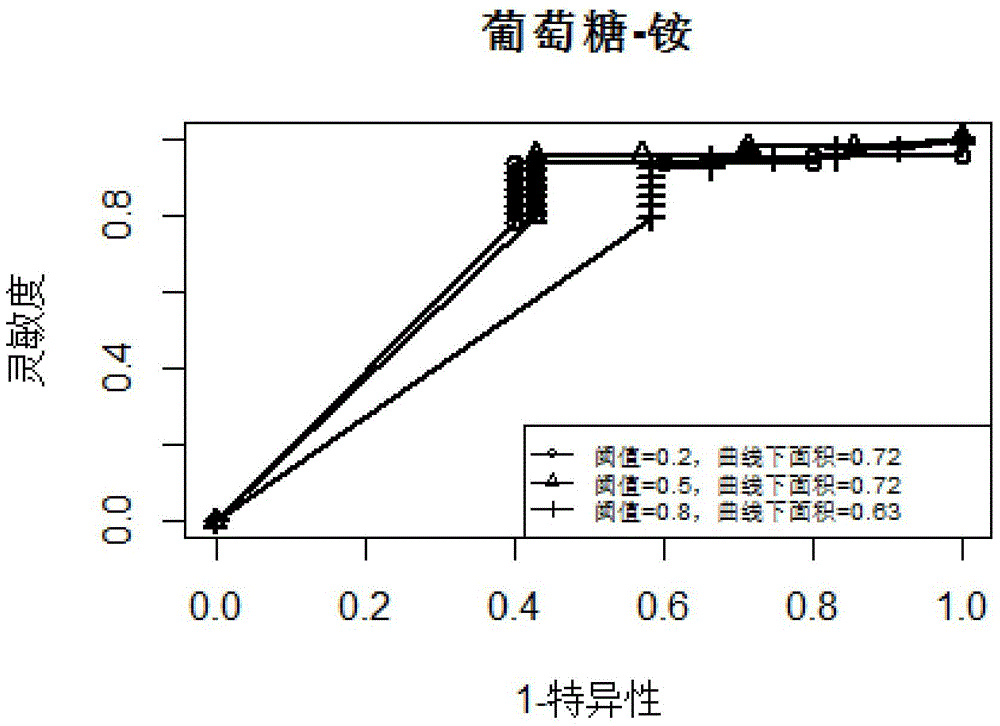
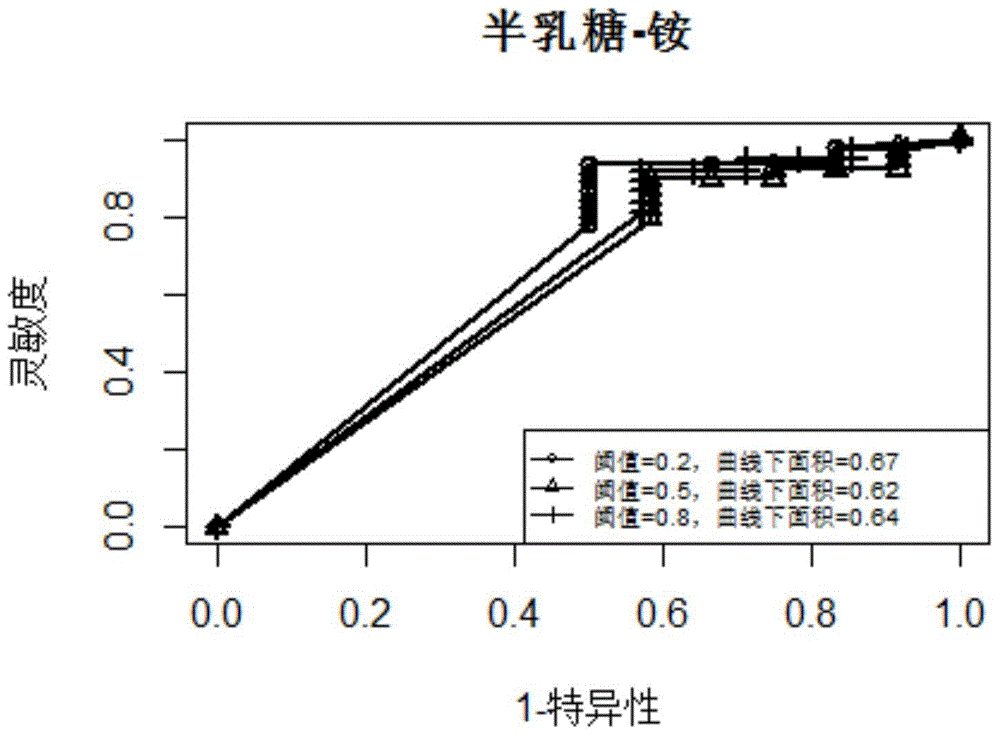
[0020] In the embodiment of the present invention, the algorithm flow chart that adopts is as follows figure 1 shown. In this embodiment, yeast is taken as an example. First, based on the collected gene expression profile data of 2929 groups of yeast, linear regression is used to infer the linear equation of the expression change of each target gene with the transcription factor. If the coefficient of a certain transcription factor in the equation is positive , it means that there is an activation effect, if the coefficient is negative, it means that there is an inhibitory effect, and if the coefficient is zero, it means that there is no regulatory effect. Then randomly select a subset of 2929 sets of expression profile data to perform 200 bootstrap linear regressions, and calculate the false discovery rate FDR according to t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com