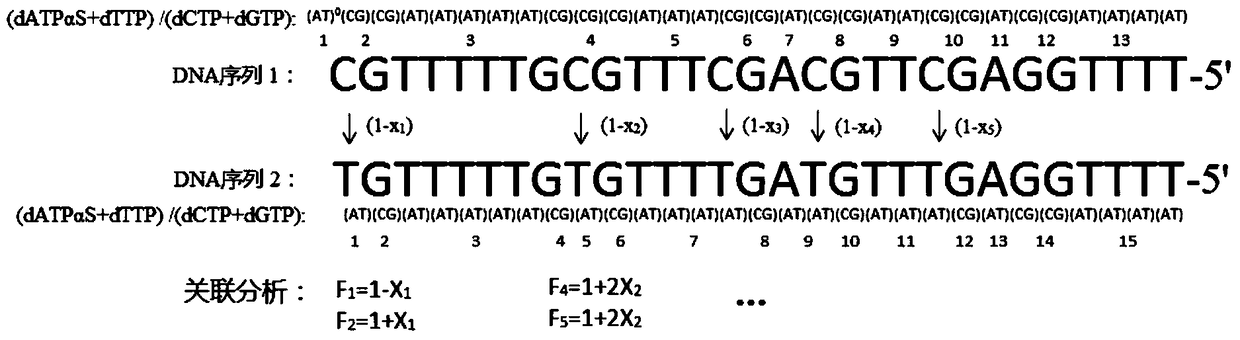A method for the quantitative detection of methylation by pyrosequencing of two nucleotide synthesis
A two-nucleotide, quantitative detection technology, applied in the biological field, can solve the problems of limited sequence analysis range, DNA template sequence analysis that cannot be multiple methylated, and exclusion, etc. Reliability, the effect of broadening the scope of analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
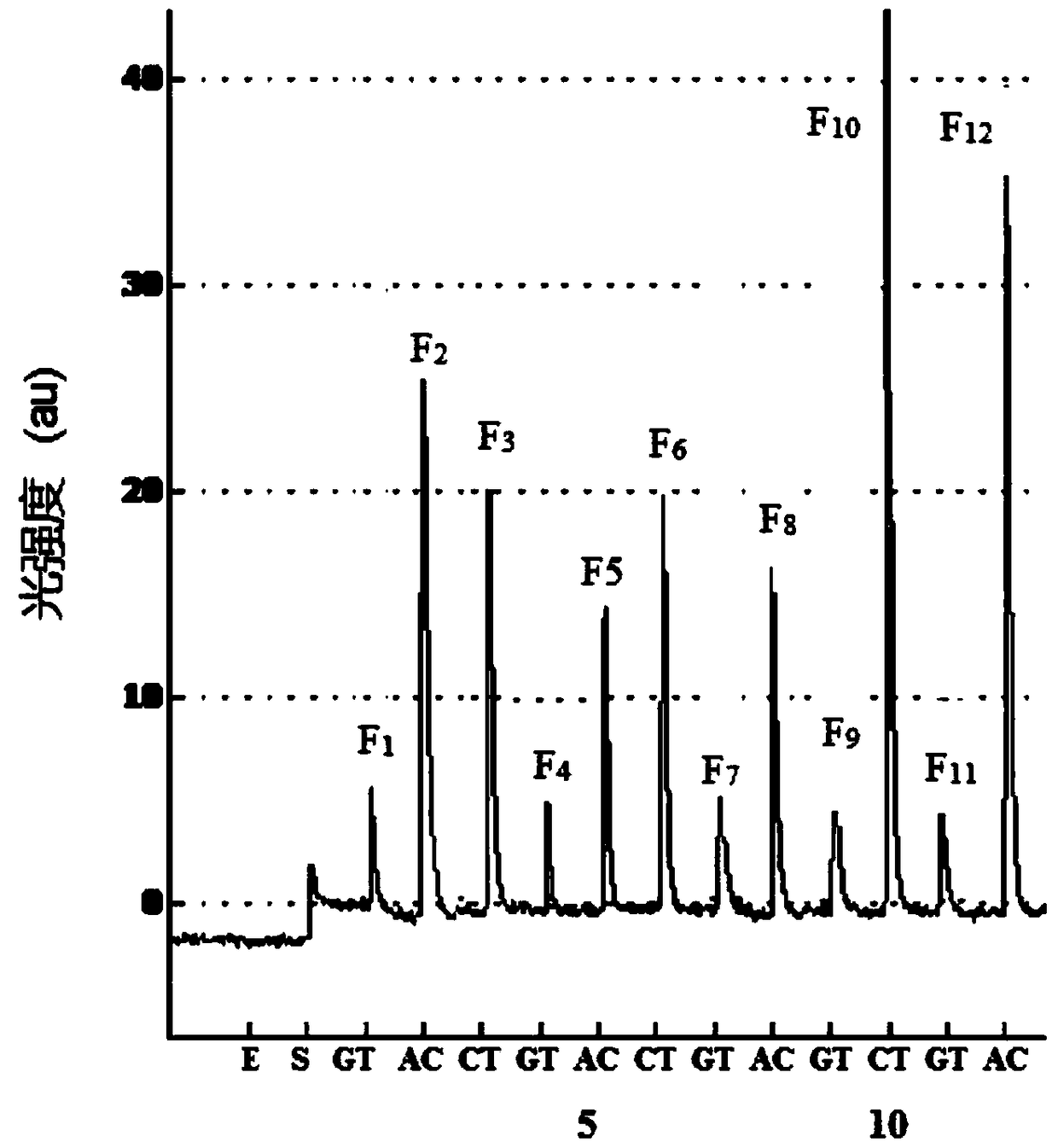
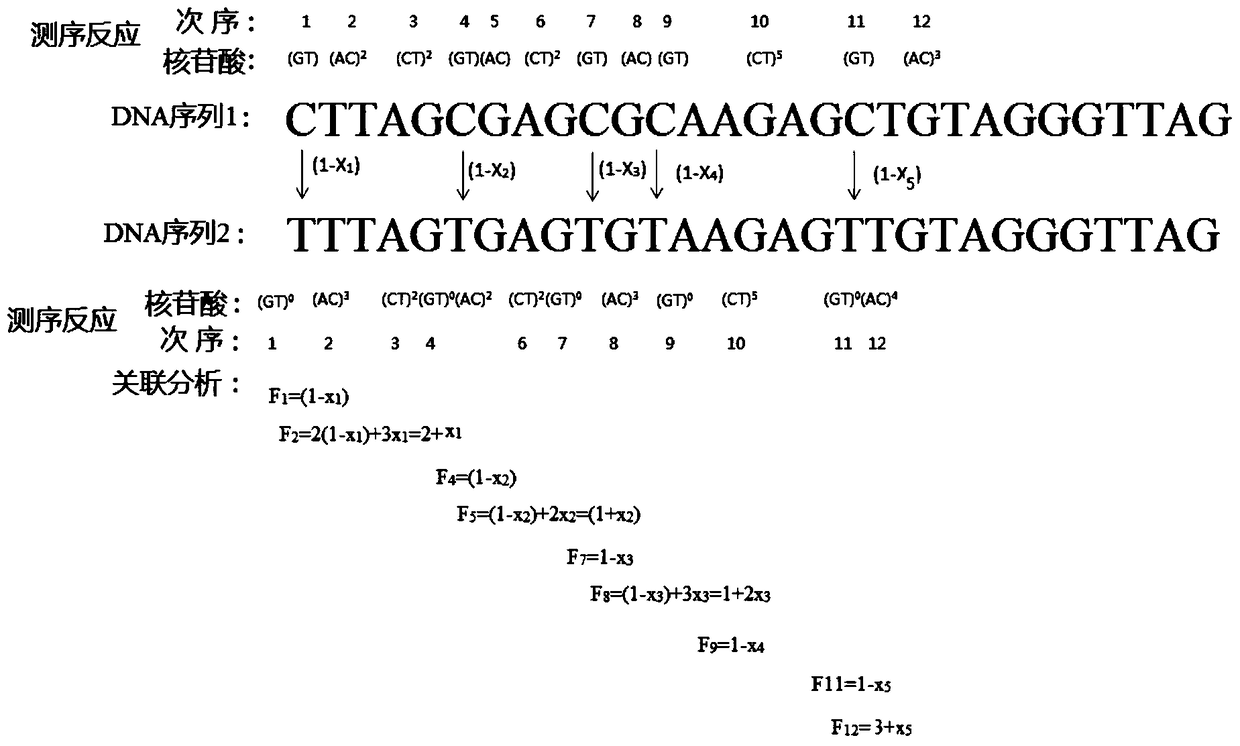
[0043] The method for analyzing the five methylation sites of the CpG island of the RARβ2 gene promoter in human samples, the specific methods include:
[0044] (1) Genomic DNA in peripheral blood was extracted from the blood sample using traditional protein kinase K and phenol / chloroform extraction methods;
[0045] (2) Genomic DNA modified by sodium bisulfite: Genomic DNA was treated with sodium bisulfite according to the operating procedure provided by CpGenome Turbo Bisulfite Modification Kit (Millipore Company), and the modified DNA was purified and recovered;
[0046] (3) PCR amplification: PCR primers: 5'-biotin-GTTGTTTGAGGATTGGGATG-3', 5'-ATACCCAAAACAAACCCTACTC-3' and 200mg of genomic DNA treated with sodium bisulfite, 0.2mM dNTP, 1UTaq DNA polymerase, 1 × Amplification buffer, 1.8mM MgCl 2 The 50 μL PCR amplification system was used for amplification. The amplification conditions were: initial denaturation at 95°C for 5 minutes; 40 thermal cycles: denaturation at 94°...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com