Double-PCR (polymerase chain reaction) method, primer and probe for identifying aquilaria sinensis
A white wood fragrance and probe technology, applied in the field of biological detection of wood species identification, can solve the problems of difficult wood identification accuracy, similar material structure, etc., and achieve the effects of protecting consumer rights, reducing interference, and less conditional dependence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Detects the primers, probes and detection kits of A. sinensis
[0039] Using resource information such as NCBI, a pair of specific amplification primers (HHPrimer-F and HHPrimer-R) was screened out, and a specific probe (HHPrimerProbe) was set in the amplification region of the primer pair. Using HHPrimer-F, HHPrimer-R and HHPrimerProbe to distinguish A. sinensis and A. yunnanensis from other species. The amplification primer pair and the probe sequence are respectively:
[0040] HHPrimer-F: 5'-CCCGTTCGCTACTTTGACTTTG-3'
[0041] HHPrimer-R: 5'-CATTACCAAGACATCATCCTCATTTTT-3'
[0042] HHPrimerProbe: 5'-VIC-TGACATAGACACAAGTC-MGB-3'.
[0043] Design a pair of specific amplification primers (BMXPrimer-F and BMXPrimer-R) and a specific probe (BMXPrimerProbe). A. sinensis was identified using BMXPrimer-F, BMXPrimer-R and BMXPrimerProbe.
[0044] BMXPrimer-F: 5'-GAGTAAGGAATACCCATTTGAATGATT-3'
[0045] BMXPrimer-R: 5'-GCGAACGGGAATTGACAGAA-3'
[0046] BMXPrimerPr...
Embodiment 2
[0070] Embodiment 2: the identification method of double qPCR
[0071] (1) Pretreatment of wood samples:
[0072] Thoroughly disinfect the wood surface with 75% ethanol, rinse with sterile water and wipe the wood dry. Cut off the surface of the wood to be tested to avoid contamination by other organizations. Take a certain amount of wood samples, add liquid nitrogen and quartz sand to a pre-cooled mortar and grind them into powder for later use. When not used immediately, the sample DNA solution was stored at -20°C until use.
[0073] (2) DNA extraction of wood samples:
[0074] The total DNA of the wood pretreated in step (1) was extracted using the Plant Genome Extraction Kit (PlantGenomicDNAKit, TIANGEN), and placed in a -40°C refrigerator for later use.
[0075] (3) The first round of qPCR amplification:
[0076] The DNA extracted in step (2) was subjected to the first round of qPCR detection. The amplification conditions are: 95°C for 10 min; 95°C for 15 s, 60°C for...
Embodiment 3
[0089] Embodiment 3: DNA extraction and detection
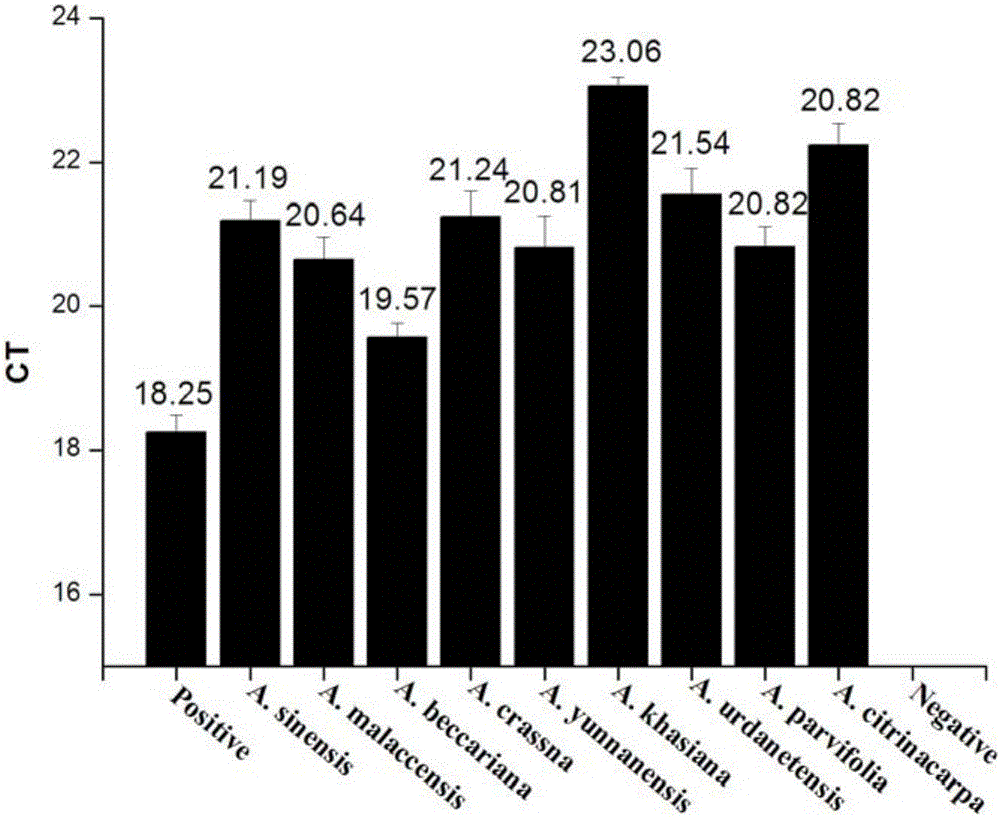
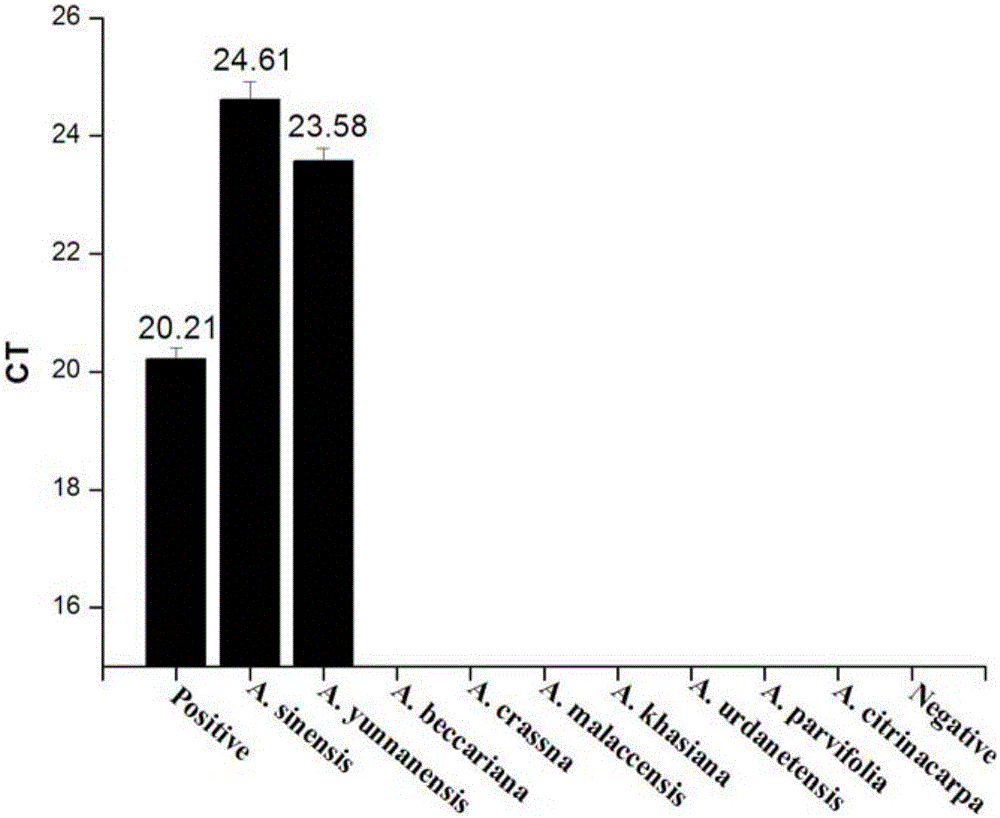
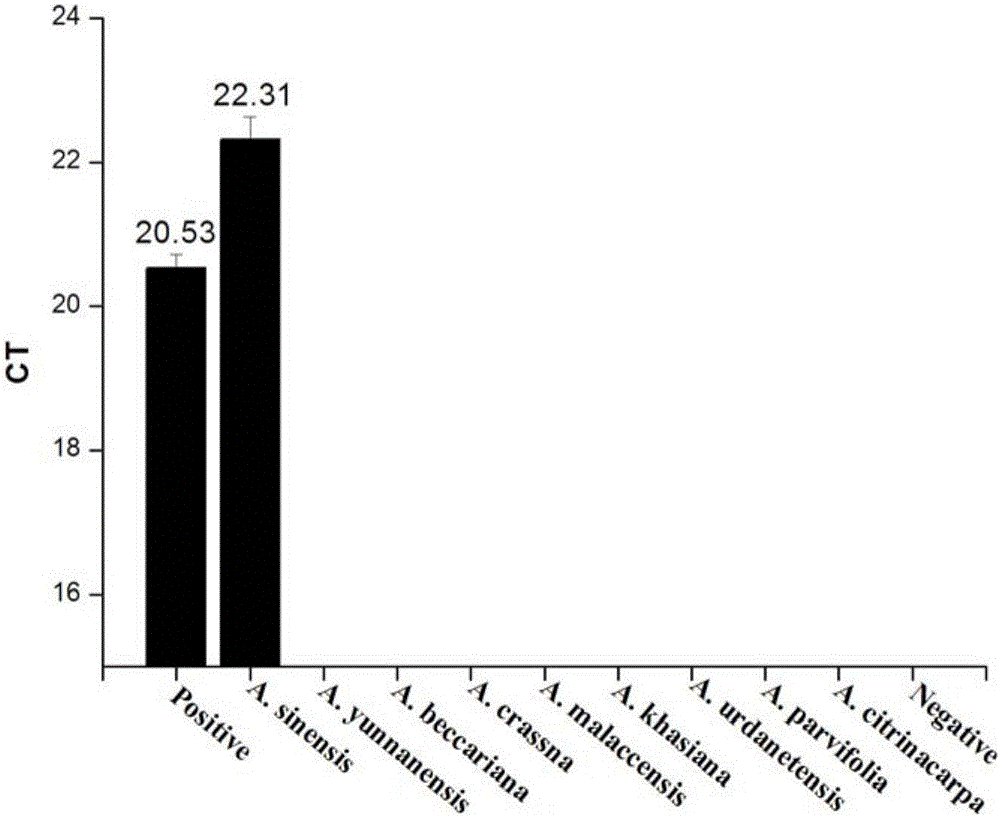
[0090] A. chinensis (experimental group 1) and the control group were extracted from A. malaccensis (Malay agarwood); A. beccariana (Northern Agarwood); A. crassna; A. urdanetensis; A. yunnanensis (Yunnan Agarwood); A. khasiana; A. parvifolia; A. citrinacarpa, a total of 8 control genomic DNAs were used as templates, and the extracted DNA was detected by using the plant endogenous gene tRNALeu and the reaction system in Example 1 and the detection method described in Example 2.
[0091] The primer and probe sequences for tRNALeu are (standard)
[0092] tRNALeu-F: 5'-CGAAATCGGTAGACGCTACG-3'
[0093] tRNALeu-R: 5'-TTCCATTGAGTCTCTGCACCT-3'
[0094] tRNALeuProbe: 5'-FAM-GCAATCCTGAGCCAAATCC-TAMRA-3'
[0095] like figure 1 It shows that the DNA of the samples of the experimental group and the control group in Example 3 have been successfully extracted.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com